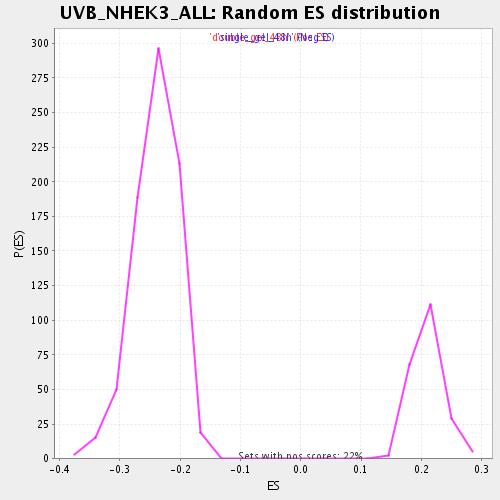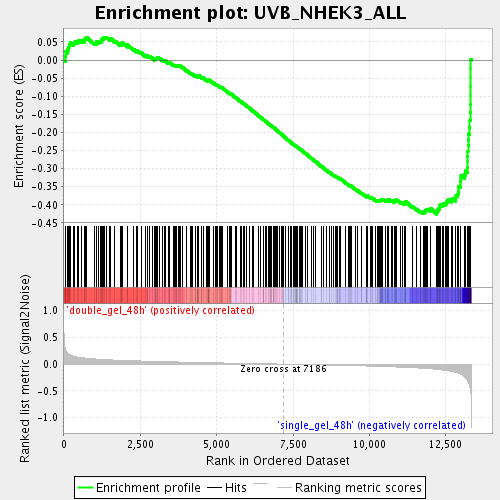
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_48h_versus_single_gel_48h.class.cls #double_gel_48h_versus_single_gel_48h_repos |
| Phenotype | class.cls#double_gel_48h_versus_single_gel_48h_repos |
| Upregulated in class | single_gel_48h |
| GeneSet | UVB_NHEK3_ALL |
| Enrichment Score (ES) | -0.42567647 |
| Normalized Enrichment Score (NES) | -1.7773732 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.046067797 |
| FWER p-Value | 0.989 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | FDFT1 | FDFT1 Entrez, Source | farnesyl-diphosphate farnesyltransferase 1 | 44 | 0.291 | 0.0109 | No |
| 2 | FASN | FASN Entrez, Source | fatty acid synthase | 60 | 0.267 | 0.0228 | No |
| 3 | GJB2 | GJB2 Entrez, Source | gap junction protein, beta 2, 26kDa (connexin 26) | 123 | 0.205 | 0.0281 | No |
| 4 | PRSS8 | PRSS8 Entrez, Source | protease, serine, 8 (prostasin) | 149 | 0.192 | 0.0356 | No |
| 5 | DHCR24 | DHCR24 Entrez, Source | 24-dehydrocholesterol reductase | 167 | 0.185 | 0.0434 | No |
| 6 | FGFBP1 | FGFBP1 Entrez, Source | fibroblast growth factor binding protein 1 | 202 | 0.172 | 0.0492 | No |
| 7 | ATF3 | ATF3 Entrez, Source | activating transcription factor 3 | 329 | 0.145 | 0.0467 | No |
| 8 | SLK | SLK Entrez, Source | STE20-like kinase (yeast) | 361 | 0.141 | 0.0512 | No |
| 9 | TFRC | TFRC Entrez, Source | transferrin receptor (p90, CD71) | 439 | 0.131 | 0.0517 | No |
| 10 | IL1RN | IL1RN Entrez, Source | interleukin 1 receptor antagonist | 475 | 0.128 | 0.0553 | No |
| 11 | ACSL1 | ACSL1 Entrez, Source | acyl-CoA synthetase long-chain family member 1 | 564 | 0.121 | 0.0544 | No |
| 12 | EPHX2 | EPHX2 Entrez, Source | epoxide hydrolase 2, cytoplasmic | 660 | 0.114 | 0.0528 | No |
| 13 | SOX9 | SOX9 Entrez, Source | SRY (sex determining region Y)-box 9 (campomelic dysplasia, autosomal sex-reversal) | 676 | 0.113 | 0.0572 | No |
| 14 | HIST2H2AA3 | HIST2H2AA3 Entrez, Source | histone cluster 2, H2aa3 | 699 | 0.112 | 0.0610 | No |
| 15 | MALL | MALL Entrez, Source | mal, T-cell differentiation protein-like | 754 | 0.109 | 0.0622 | No |
| 16 | GDF15 | GDF15 Entrez, Source | growth differentiation factor 15 | 1016 | 0.097 | 0.0469 | No |
| 17 | MCFD2 | MCFD2 Entrez, Source | multiple coagulation factor deficiency 2 | 1070 | 0.095 | 0.0475 | No |
| 18 | PBEF1 | PBEF1 Entrez, Source | pre-B-cell colony enhancing factor 1 | 1074 | 0.094 | 0.0519 | No |
| 19 | UBE2I | UBE2I Entrez, Source | ubiquitin-conjugating enzyme E2I (UBC9 homolog, yeast) | 1146 | 0.092 | 0.0509 | No |
| 20 | ITGB5 | ITGB5 Entrez, Source | integrin, beta 5 | 1194 | 0.090 | 0.0517 | No |
| 21 | DUSP4 | DUSP4 Entrez, Source | dual specificity phosphatase 4 | 1225 | 0.089 | 0.0538 | No |
| 22 | GCLC | GCLC Entrez, Source | glutamate-cysteine ligase, catalytic subunit | 1237 | 0.088 | 0.0573 | No |
| 23 | PIM1 | PIM1 Entrez, Source | pim-1 oncogene | 1264 | 0.087 | 0.0595 | No |
| 24 | IER2 | IER2 Entrez, Source | immediate early response 2 | 1296 | 0.086 | 0.0614 | No |
| 25 | DSC2 | DSC2 Entrez, Source | desmocollin 2 | 1330 | 0.085 | 0.0630 | No |
| 26 | MTHFD1 | MTHFD1 Entrez, Source | methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase | 1392 | 0.083 | 0.0624 | No |
| 27 | ME1 | ME1 Entrez, Source | malic enzyme 1, NADP(+)-dependent, cytosolic | 1493 | 0.080 | 0.0586 | No |
| 28 | HMGA2 | HMGA2 Entrez, Source | high mobility group AT-hook 2 | 1533 | 0.079 | 0.0595 | No |
| 29 | UBE2D2 | UBE2D2 Entrez, Source | ubiquitin-conjugating enzyme E2D 2 (UBC4/5 homolog, yeast) | 1670 | 0.075 | 0.0528 | No |
| 30 | UBE2V2 | UBE2V2 Entrez, Source | ubiquitin-conjugating enzyme E2 variant 2 | 1845 | 0.071 | 0.0429 | No |
| 31 | VEGF | VEGF Entrez, Source | vascular endothelial growth factor | 1883 | 0.070 | 0.0435 | No |
| 32 | MYC | MYC Entrez, Source | v-myc myelocytomatosis viral oncogene homolog (avian) | 1896 | 0.070 | 0.0461 | No |
| 33 | YARS | YARS Entrez, Source | tyrosyl-tRNA synthetase | 1907 | 0.070 | 0.0487 | No |
| 34 | EPRS | EPRS Entrez, Source | glutamyl-prolyl-tRNA synthetase | 2076 | 0.066 | 0.0391 | No |
| 35 | SP3 | SP3 Entrez, Source | Sp3 transcription factor | 2081 | 0.066 | 0.0420 | No |
| 36 | RAF1 | RAF1 Entrez, Source | v-raf-1 murine leukemia viral oncogene homolog 1 | 2284 | 0.062 | 0.0296 | No |
| 37 | ADK | ADK Entrez, Source | adenosine kinase | 2387 | 0.061 | 0.0248 | No |
| 38 | SLC7A5 | SLC7A5 Entrez, Source | solute carrier family 7 (cationic amino acid transporter, y+ system), member 5 | 2396 | 0.060 | 0.0271 | No |
| 39 | CD9 | CD9 Entrez, Source | CD9 molecule | 2527 | 0.058 | 0.0200 | No |
| 40 | LIF | LIF Entrez, Source | leukemia inhibitory factor (cholinergic differentiation factor) | 2660 | 0.056 | 0.0127 | No |
| 41 | ACY1 | ACY1 Entrez, Source | aminoacylase 1 | 2722 | 0.055 | 0.0107 | No |
| 42 | HMOX1 | HMOX1 Entrez, Source | heme oxygenase (decycling) 1 | 2730 | 0.055 | 0.0128 | No |
| 43 | PTP4A1 | PTP4A1 Entrez, Source | protein tyrosine phosphatase type IVA, member 1 | 2810 | 0.053 | 0.0094 | No |
| 44 | ATP5O | ATP5O Entrez, Source | ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit (oligomycin sensitivity conferring protein) | 2883 | 0.052 | 0.0064 | No |
| 45 | HNRPA1 | HNRPA1 Entrez, Source | heterogeneous nuclear ribonucleoprotein A1 | 2977 | 0.050 | 0.0017 | No |
| 46 | TNFRSF1A | TNFRSF1A Entrez, Source | tumor necrosis factor receptor superfamily, member 1A | 2994 | 0.050 | 0.0030 | No |
| 47 | PTGS1 | PTGS1 Entrez, Source | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | 3007 | 0.050 | 0.0045 | No |
| 48 | R3HDM1 | R3HDM1 Entrez, Source | R3H domain containing 1 | 3028 | 0.050 | 0.0054 | No |
| 49 | PPIF | PPIF Entrez, Source | peptidylprolyl isomerase F (cyclophilin F) | 3047 | 0.049 | 0.0064 | No |
| 50 | ASS1 | ASS1 Entrez, Source | argininosuccinate synthetase 1 | 3063 | 0.049 | 0.0077 | No |
| 51 | AARS | AARS Entrez, Source | alanyl-tRNA synthetase | 3123 | 0.048 | 0.0055 | No |
| 52 | FHL1 | FHL1 Entrez, Source | four and a half LIM domains 1 | 3226 | 0.047 | 0.0000 | No |
| 53 | USP10 | USP10 Entrez, Source | ubiquitin specific peptidase 10 | 3286 | 0.046 | -0.0022 | No |
| 54 | OAS1 | OAS1 Entrez, Source | 2',5'-oligoadenylate synthetase 1, 40/46kDa | 3313 | 0.046 | -0.0020 | No |
| 55 | LAMA3 | LAMA3 Entrez, Source | laminin, alpha 3 | 3419 | 0.044 | -0.0079 | No |
| 56 | WWP1 | WWP1 Entrez, Source | WW domain containing E3 ubiquitin protein ligase 1 | 3459 | 0.044 | -0.0087 | No |
| 57 | RRAGA | RRAGA Entrez, Source | Ras-related GTP binding A | 3466 | 0.043 | -0.0071 | No |
| 58 | ATF4 | ATF4 Entrez, Source | activating transcription factor 4 (tax-responsive enhancer element B67) | 3579 | 0.042 | -0.0136 | No |
| 59 | MAPK6 | MAPK6 Entrez, Source | mitogen-activated protein kinase 6 | 3619 | 0.042 | -0.0145 | No |
| 60 | POLD2 | POLD2 Entrez, Source | polymerase (DNA directed), delta 2, regulatory subunit 50kDa | 3642 | 0.041 | -0.0142 | No |
| 61 | ETF1 | ETF1 Entrez, Source | eukaryotic translation termination factor 1 | 3680 | 0.041 | -0.0150 | No |
| 62 | CLINT1 | CLINT1 Entrez, Source | clathrin interactor 1 | 3693 | 0.041 | -0.0140 | No |
| 63 | MYST3 | MYST3 Entrez, Source | MYST histone acetyltransferase (monocytic leukemia) 3 | 3748 | 0.040 | -0.0162 | No |
| 64 | YY1 | YY1 Entrez, Source | YY1 transcription factor | 3788 | 0.039 | -0.0172 | No |
| 65 | ACO2 | ACO2 Entrez, Source | aconitase 2, mitochondrial | 3799 | 0.039 | -0.0161 | No |
| 66 | VSNL1 | VSNL1 Entrez, Source | visinin-like 1 | 3879 | 0.038 | -0.0202 | No |
| 67 | FLNB | FLNB Entrez, Source | filamin B, beta (actin binding protein 278) | 4024 | 0.036 | -0.0295 | No |
| 68 | SAFB | SAFB Entrez, Source | scaffold attachment factor B | 4133 | 0.035 | -0.0361 | No |
| 69 | RSN | RSN Entrez, Source | restin (Reed-Steinberg cell-expressed intermediate filament-associated protein) | 4173 | 0.034 | -0.0374 | No |
| 70 | CEBPD | CEBPD Entrez, Source | CCAAT/enhancer binding protein (C/EBP), delta | 4223 | 0.034 | -0.0395 | No |
| 71 | CDKN3 | CDKN3 Entrez, Source | cyclin-dependent kinase inhibitor 3 (CDK2-associated dual specificity phosphatase) | 4303 | 0.033 | -0.0439 | No |
| 72 | PLA2G4A | PLA2G4A Entrez, Source | phospholipase A2, group IVA (cytosolic, calcium-dependent) | 4307 | 0.033 | -0.0426 | No |
| 73 | CELSR2 | CELSR2 Entrez, Source | cadherin, EGF LAG seven-pass G-type receptor 2 (flamingo homolog, Drosophila) | 4367 | 0.032 | -0.0455 | No |
| 74 | HSD17B4 | HSD17B4 Entrez, Source | hydroxysteroid (17-beta) dehydrogenase 4 | 4370 | 0.032 | -0.0441 | No |
| 75 | DDX1 | DDX1 Entrez, Source | DEAD (Asp-Glu-Ala-Asp) box polypeptide 1 | 4387 | 0.031 | -0.0438 | No |
| 76 | CALB2 | CALB2 Entrez, Source | calbindin 2, 29kDa (calretinin) | 4390 | 0.031 | -0.0424 | No |
| 77 | ANKRD15 | ANKRD15 Entrez, Source | ankyrin repeat domain 15 | 4401 | 0.031 | -0.0417 | No |
| 78 | EIF2B1 | EIF2B1 Entrez, Source | eukaryotic translation initiation factor 2B, subunit 1 alpha, 26kDa | 4517 | 0.030 | -0.0490 | No |
| 79 | RBP1 | RBP1 Entrez, Source | retinol binding protein 1, cellular | 4518 | 0.030 | -0.0475 | No |
| 80 | SDHA | SDHA Entrez, Source | succinate dehydrogenase complex, subunit A, flavoprotein (Fp) | 4559 | 0.029 | -0.0492 | No |
| 81 | EIF2S1 | EIF2S1 Entrez, Source | eukaryotic translation initiation factor 2, subunit 1 alpha, 35kDa | 4657 | 0.028 | -0.0552 | No |
| 82 | THBS2 | THBS2 Entrez, Source | thrombospondin 2 | 4709 | 0.027 | -0.0578 | No |
| 83 | BRD2 | BRD2 Entrez, Source | bromodomain containing 2 | 4720 | 0.027 | -0.0572 | No |
| 84 | DDX39 | DDX39 Entrez, Source | DEAD (Asp-Glu-Ala-Asp) box polypeptide 39 | 4724 | 0.027 | -0.0561 | No |
| 85 | EIF4G2 | EIF4G2 Entrez, Source | eukaryotic translation initiation factor 4 gamma, 2 | 4729 | 0.027 | -0.0551 | No |
| 86 | XRCC6 | XRCC6 Entrez, Source | X-ray repair complementing defective repair in Chinese hamster cells 6 (Ku autoantigen, 70kDa) | 4746 | 0.027 | -0.0550 | No |
| 87 | IL4R | IL4R Entrez, Source | interleukin 4 receptor | 4765 | 0.027 | -0.0550 | No |
| 88 | JUND | JUND Entrez, Source | jun D proto-oncogene | 4884 | 0.025 | -0.0628 | No |
| 89 | GALNT3 | GALNT3 Entrez, Source | UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 3 (GalNAc-T3) | 4949 | 0.024 | -0.0665 | No |
| 90 | PTPN12 | PTPN12 Entrez, Source | protein tyrosine phosphatase, non-receptor type 12 | 5000 | 0.024 | -0.0692 | No |
| 91 | RAD23B | RAD23B Entrez, Source | RAD23 homolog B (S. cerevisiae) | 5021 | 0.024 | -0.0696 | No |
| 92 | MMP9 | MMP9 Entrez, Source | matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) | 5075 | 0.023 | -0.0725 | No |
| 93 | DUSP1 | DUSP1 Entrez, Source | dual specificity phosphatase 1 | 5117 | 0.022 | -0.0746 | No |
| 94 | ARL4A | ARL4A Entrez, Source | ADP-ribosylation factor-like 4A | 5144 | 0.022 | -0.0755 | No |
| 95 | PYGL | PYGL Entrez, Source | phosphorylase, glycogen; liver (Hers disease, glycogen storage disease type VI) | 5160 | 0.022 | -0.0755 | No |
| 96 | GADD45A | GADD45A Entrez, Source | growth arrest and DNA-damage-inducible, alpha | 5178 | 0.022 | -0.0758 | No |
| 97 | COPS8 | COPS8 Entrez, Source | COP9 constitutive photomorphogenic homolog subunit 8 (Arabidopsis) | 5364 | 0.020 | -0.0890 | No |
| 98 | XPO1 | XPO1 Entrez, Source | exportin 1 (CRM1 homolog, yeast) | 5406 | 0.019 | -0.0912 | No |
| 99 | SON | SON Entrez, Source | SON DNA binding protein | 5446 | 0.019 | -0.0932 | No |
| 100 | VDAC2 | VDAC2 Entrez, Source | voltage-dependent anion channel 2 | 5456 | 0.019 | -0.0930 | No |
| 101 | PSMA6 | PSMA6 Entrez, Source | proteasome (prosome, macropain) subunit, alpha type, 6 | 5475 | 0.019 | -0.0935 | No |
| 102 | XBP1 | XBP1 Entrez, Source | X-box binding protein 1 | 5608 | 0.017 | -0.1027 | No |
| 103 | GSK3A | GSK3A Entrez, Source | glycogen synthase kinase 3 alpha | 5648 | 0.017 | -0.1049 | No |
| 104 | CCND2 | CCND2 Entrez, Source | cyclin D2 | 5771 | 0.015 | -0.1135 | No |
| 105 | CDKN1A | CDKN1A Entrez, Source | cyclin-dependent kinase inhibitor 1A (p21, Cip1) | 5802 | 0.015 | -0.1151 | No |
| 106 | BAMBI | BAMBI Entrez, Source | BMP and activin membrane-bound inhibitor homolog (Xenopus laevis) | 5874 | 0.014 | -0.1198 | No |
| 107 | HK1 | HK1 Entrez, Source | hexokinase 1 | 5904 | 0.014 | -0.1214 | No |
| 108 | GSPT1 | GSPT1 Entrez, Source | G1 to S phase transition 1 | 5981 | 0.013 | -0.1265 | No |
| 109 | TIAM1 | TIAM1 Entrez, Source | T-cell lymphoma invasion and metastasis 1 | 6064 | 0.012 | -0.1322 | No |
| 110 | RPS24 | RPS24 Entrez, Source | ribosomal protein S24 | 6181 | 0.011 | -0.1406 | No |
| 111 | HMGB1 | HMGB1 Entrez, Source | high-mobility group box 1 | 6219 | 0.011 | -0.1429 | No |
| 112 | EXOSC7 | EXOSC7 Entrez, Source | exosome component 7 | 6360 | 0.009 | -0.1531 | No |
| 113 | FGFR3 | FGFR3 Entrez, Source | fibroblast growth factor receptor 3 (achondroplasia, thanatophoric dwarfism) | 6436 | 0.008 | -0.1585 | No |
| 114 | PPIB | PPIB Entrez, Source | peptidylprolyl isomerase B (cyclophilin B) | 6516 | 0.007 | -0.1642 | No |
| 115 | RNF4 | RNF4 Entrez, Source | ring finger protein 4 | 6533 | 0.007 | -0.1650 | No |
| 116 | PGRMC1 | PGRMC1 Entrez, Source | progesterone receptor membrane component 1 | 6542 | 0.007 | -0.1653 | No |
| 117 | PKN2 | PKN2 Entrez, Source | protein kinase N2 | 6600 | 0.006 | -0.1693 | No |
| 118 | TSG101 | TSG101 Entrez, Source | tumor susceptibility gene 101 | 6620 | 0.006 | -0.1705 | No |
| 119 | UBXD2 | UBXD2 Entrez, Source | UBX domain containing 2 | 6708 | 0.005 | -0.1769 | No |
| 120 | PON2 | PON2 Entrez, Source | paraoxonase 2 | 6716 | 0.005 | -0.1772 | No |
| 121 | ZNF148 | ZNF148 Entrez, Source | zinc finger protein 148 | 6751 | 0.005 | -0.1795 | No |
| 122 | SAP18 | SAP18 Entrez, Source | Sin3A-associated protein, 18kDa | 6775 | 0.004 | -0.1811 | No |
| 123 | ATP6V1B2 | ATP6V1B2 Entrez, Source | ATPase, H+ transporting, lysosomal 56/58kDa, V1 subunit B2 | 6788 | 0.004 | -0.1818 | No |
| 124 | CTSC | CTSC Entrez, Source | cathepsin C | 6806 | 0.004 | -0.1829 | No |
| 125 | PREP | PREP Entrez, Source | prolyl endopeptidase | 6861 | 0.004 | -0.1869 | No |
| 126 | AKR1A1 | AKR1A1 Entrez, Source | aldo-keto reductase family 1, member A1 (aldehyde reductase) | 6869 | 0.003 | -0.1872 | No |
| 127 | HSPH1 | HSPH1 Entrez, Source | heat shock 105kDa/110kDa protein 1 | 6870 | 0.003 | -0.1871 | No |
| 128 | CDC20 | CDC20 Entrez, Source | CDC20 cell division cycle 20 homolog (S. cerevisiae) | 6894 | 0.003 | -0.1887 | No |
| 129 | LTB4DH | LTB4DH Entrez, Source | leukotriene B4 12-hydroxydehydrogenase | 6909 | 0.003 | -0.1896 | No |
| 130 | GPX2 | GPX2 Entrez, Source | glutathione peroxidase 2 (gastrointestinal) | 6946 | 0.003 | -0.1922 | No |
| 131 | CSNK2A1 | CSNK2A1 Entrez, Source | casein kinase 2, alpha 1 polypeptide | 6997 | 0.002 | -0.1960 | No |
| 132 | KPNA2 | KPNA2 Entrez, Source | karyopherin alpha 2 (RAG cohort 1, importin alpha 1) | 7056 | 0.001 | -0.2003 | No |
| 133 | LCN2 | LCN2 Entrez, Source | lipocalin 2 (oncogene 24p3) | 7114 | 0.001 | -0.2047 | No |
| 134 | TMED10 | TMED10 Entrez, Source | transmembrane emp24-like trafficking protein 10 (yeast) | 7149 | 0.000 | -0.2072 | No |
| 135 | ATP6V0D1 | ATP6V0D1 Entrez, Source | ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d1 | 7193 | -0.000 | -0.2105 | No |
| 136 | PSMA5 | PSMA5 Entrez, Source | proteasome (prosome, macropain) subunit, alpha type, 5 | 7199 | -0.000 | -0.2109 | No |
| 137 | BPGM | BPGM Entrez, Source | 2,3-bisphosphoglycerate mutase | 7240 | -0.001 | -0.2139 | No |
| 138 | PGM1 | PGM1 Entrez, Source | phosphoglucomutase 1 | 7335 | -0.002 | -0.2210 | No |
| 139 | HNRPU | HNRPU Entrez, Source | heterogeneous nuclear ribonucleoprotein U (scaffold attachment factor A) | 7350 | -0.002 | -0.2220 | No |
| 140 | CYB5A | CYB5A Entrez, Source | cytochrome b5 type A (microsomal) | 7424 | -0.003 | -0.2275 | No |
| 141 | KCNK1 | KCNK1 Entrez, Source | potassium channel, subfamily K, member 1 | 7451 | -0.003 | -0.2293 | No |
| 142 | PTPRK | PTPRK Entrez, Source | protein tyrosine phosphatase, receptor type, K | 7463 | -0.003 | -0.2300 | No |
| 143 | BLVRA | BLVRA Entrez, Source | biliverdin reductase A | 7498 | -0.004 | -0.2324 | No |
| 144 | PSMA4 | PSMA4 Entrez, Source | proteasome (prosome, macropain) subunit, alpha type, 4 | 7508 | -0.004 | -0.2329 | No |
| 145 | PPP2R2A | PPP2R2A Entrez, Source | protein phosphatase 2 (formerly 2A), regulatory subunit B (PR 52), alpha isoform | 7540 | -0.004 | -0.2351 | No |
| 146 | STK25 | STK25 Entrez, Source | serine/threonine kinase 25 (STE20 homolog, yeast) | 7573 | -0.005 | -0.2373 | No |
| 147 | PSMC6 | PSMC6 Entrez, Source | proteasome (prosome, macropain) 26S subunit, ATPase, 6 | 7584 | -0.005 | -0.2378 | No |
| 148 | ASNS | ASNS Entrez, Source | asparagine synthetase | 7614 | -0.005 | -0.2398 | No |
| 149 | RASA1 | RASA1 Entrez, Source | RAS p21 protein activator (GTPase activating protein) 1 | 7619 | -0.005 | -0.2399 | No |
| 150 | RPL3 | RPL3 Entrez, Source | ribosomal protein L3 | 7626 | -0.005 | -0.2401 | No |
| 151 | COPS2 | COPS2 Entrez, Source | COP9 constitutive photomorphogenic homolog subunit 2 (Arabidopsis) | 7649 | -0.005 | -0.2415 | No |
| 152 | TOMM20 | TOMM20 Entrez, Source | translocase of outer mitochondrial membrane 20 homolog (yeast) | 7655 | -0.005 | -0.2416 | No |
| 153 | ATP1B1 | ATP1B1 Entrez, Source | ATPase, Na+/K+ transporting, beta 1 polypeptide | 7708 | -0.006 | -0.2453 | No |
| 154 | PSMD6 | PSMD6 Entrez, Source | proteasome (prosome, macropain) 26S subunit, non-ATPase, 6 | 7715 | -0.006 | -0.2455 | No |
| 155 | TCEB1 | TCEB1 Entrez, Source | transcription elongation factor B (SIII), polypeptide 1 (15kDa, elongin C) | 7731 | -0.006 | -0.2463 | No |
| 156 | ST6GALNAC2 | ST6GALNAC2 Entrez, Source | ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 2 | 7740 | -0.006 | -0.2466 | No |
| 157 | PSMC1 | PSMC1 Entrez, Source | proteasome (prosome, macropain) 26S subunit, ATPase, 1 | 7772 | -0.007 | -0.2487 | No |
| 158 | GLG1 | GLG1 Entrez, Source | golgi apparatus protein 1 | 7800 | -0.007 | -0.2504 | No |
| 159 | HNRPK | HNRPK Entrez, Source | heterogeneous nuclear ribonucleoprotein K | 7921 | -0.009 | -0.2592 | No |
| 160 | ARL6IP1 | ARL6IP1 Entrez, Source | ADP-ribosylation factor-like 6 interacting protein 1 | 7958 | -0.009 | -0.2615 | No |
| 161 | COL5A2 | COL5A2 Entrez, Source | collagen, type V, alpha 2 | 7962 | -0.009 | -0.2613 | No |
| 162 | MAD2L1BP | MAD2L1BP Entrez, Source | MAD2L1 binding protein | 8118 | -0.011 | -0.2726 | No |
| 163 | PSME4 | PSME4 Entrez, Source | proteasome (prosome, macropain) activator subunit 4 | 8172 | -0.011 | -0.2761 | No |
| 164 | FBN2 | FBN2 Entrez, Source | fibrillin 2 (congenital contractural arachnodactyly) | 8235 | -0.012 | -0.2803 | No |
| 165 | PSMC2 | PSMC2 Entrez, Source | proteasome (prosome, macropain) 26S subunit, ATPase, 2 | 8416 | -0.014 | -0.2933 | No |
| 166 | DUSP5 | DUSP5 Entrez, Source | dual specificity phosphatase 5 | 8419 | -0.014 | -0.2928 | No |
| 167 | PRDX6 | PRDX6 Entrez, Source | peroxiredoxin 6 | 8502 | -0.016 | -0.2983 | No |
| 168 | DAZAP2 | DAZAP2 Entrez, Source | DAZ associated protein 2 | 8593 | -0.017 | -0.3044 | No |
| 169 | BNIP3 | BNIP3 Entrez, Source | BCL2/adenovirus E1B 19kDa interacting protein 3 | 8690 | -0.018 | -0.3108 | No |
| 170 | RAB2 | RAB2 Entrez, Source | RAB2, member RAS oncogene family | 8693 | -0.018 | -0.3101 | No |
| 171 | HSP90AA1 | HSP90AA1 Entrez, Source | heat shock protein 90kDa alpha (cytosolic), class A member 1 | 8764 | -0.019 | -0.3145 | No |
| 172 | UBE2G1 | UBE2G1 Entrez, Source | ubiquitin-conjugating enzyme E2G 1 (UBC7 homolog, yeast) | 8829 | -0.020 | -0.3185 | No |
| 173 | ATIC | ATIC Entrez, Source | 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase | 8886 | -0.020 | -0.3218 | No |
| 174 | PPP1CC | PPP1CC Entrez, Source | protein phosphatase 1, catalytic subunit, gamma isoform | 8913 | -0.021 | -0.3228 | No |
| 175 | PFN2 | PFN2 Entrez, Source | profilin 2 | 8946 | -0.021 | -0.3242 | No |
| 176 | PPP3CA | PPP3CA Entrez, Source | protein phosphatase 3 (formerly 2B), catalytic subunit, alpha isoform (calcineurin A alpha) | 8966 | -0.021 | -0.3246 | No |
| 177 | CTR9 | CTR9 Entrez, Source | Ctr9, Paf1/RNA polymerase II complex component, homolog (S. cerevisiae) | 9023 | -0.022 | -0.3278 | No |
| 178 | CDK7 | CDK7 Entrez, Source | cyclin-dependent kinase 7 (MO15 homolog, Xenopus laevis, cdk-activating kinase) | 9024 | -0.022 | -0.3267 | No |
| 179 | STAT1 | STAT1 Entrez, Source | signal transducer and activator of transcription 1, 91kDa | 9067 | -0.023 | -0.3288 | No |
| 180 | TAF9 | TAF9 Entrez, Source | TAF9 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 32kDa | 9217 | -0.025 | -0.3390 | No |
| 181 | ALDH9A1 | ALDH9A1 Entrez, Source | aldehyde dehydrogenase 9 family, member A1 | 9309 | -0.026 | -0.3447 | No |
| 182 | NFE2L2 | NFE2L2 Entrez, Source | nuclear factor (erythroid-derived 2)-like 2 | 9356 | -0.027 | -0.3469 | No |
| 183 | CNN3 | CNN3 Entrez, Source | calponin 3, acidic | 9383 | -0.027 | -0.3475 | No |
| 184 | DNAJA1 | DNAJA1 Entrez, Source | DnaJ (Hsp40) homolog, subfamily A, member 1 | 9406 | -0.028 | -0.3479 | No |
| 185 | ATP6V0C | ATP6V0C Entrez, Source | ATPase, H+ transporting, lysosomal 16kDa, V0 subunit c | 9541 | -0.030 | -0.3567 | No |
| 186 | ATP1B3 | ATP1B3 Entrez, Source | ATPase, Na+/K+ transporting, beta 3 polypeptide | 9618 | -0.031 | -0.3610 | No |
| 187 | GLUL | GLUL Entrez, Source | glutamate-ammonia ligase (glutamine synthetase) | 9734 | -0.033 | -0.3682 | No |
| 188 | RND3 | RND3 Entrez, Source | Rho family GTPase 3 | 9751 | -0.033 | -0.3678 | No |
| 189 | MMP10 | MMP10 Entrez, Source | matrix metallopeptidase 10 (stromelysin 2) | 9894 | -0.035 | -0.3770 | No |
| 190 | DUT | DUT Entrez, Source | dUTP pyrophosphatase | 9896 | -0.035 | -0.3754 | No |
| 191 | NMT1 | NMT1 Entrez, Source | N-myristoyltransferase 1 | 9908 | -0.035 | -0.3745 | No |
| 192 | ITGB4 | ITGB4 Entrez, Source | integrin, beta 4 | 9935 | -0.035 | -0.3748 | No |
| 193 | WBSCR1 | WBSCR1 Entrez, Source | Williams-Beuren syndrome chromosome region 1 | 10043 | -0.037 | -0.3811 | No |
| 194 | PCMT1 | PCMT1 Entrez, Source | protein-L-isoaspartate (D-aspartate) O-methyltransferase | 10054 | -0.038 | -0.3800 | No |
| 195 | GPIAP1 | GPIAP1 Entrez, Source | GPI-anchored membrane protein 1 | 10100 | -0.038 | -0.3816 | No |
| 196 | MYD88 | MYD88 Entrez, Source | myeloid differentiation primary response gene (88) | 10202 | -0.040 | -0.3874 | No |
| 197 | SLC39A6 | SLC39A6 Entrez, Source | solute carrier family 39 (zinc transporter), member 6 | 10251 | -0.041 | -0.3891 | No |
| 198 | RAB21 | RAB21 Entrez, Source | RAB21, member RAS oncogene family | 10253 | -0.041 | -0.3871 | No |
| 199 | CCNG1 | CCNG1 Entrez, Source | cyclin G1 | 10280 | -0.041 | -0.3871 | No |
| 200 | SEPT2 | SEPT2 Entrez, Source | septin 2 | 10339 | -0.042 | -0.3895 | No |
| 201 | PPP2CB | PPP2CB Entrez, Source | protein phosphatase 2 (formerly 2A), catalytic subunit, beta isoform | 10348 | -0.042 | -0.3881 | No |
| 202 | GTF2B | GTF2B Entrez, Source | general transcription factor IIB | 10363 | -0.043 | -0.3870 | No |
| 203 | NFKBIA | NFKBIA Entrez, Source | nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha | 10388 | -0.043 | -0.3868 | No |
| 204 | TRAM1 | TRAM1 Entrez, Source | translocation associated membrane protein 1 | 10392 | -0.043 | -0.3849 | No |
| 205 | GTF2A2 | GTF2A2 Entrez, Source | general transcription factor IIA, 2, 12kDa | 10421 | -0.044 | -0.3849 | No |
| 206 | IER3 | IER3 Entrez, Source | immediate early response 3 | 10439 | -0.044 | -0.3840 | No |
| 207 | ITGB1 | ITGB1 Entrez, Source | integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) | 10521 | -0.046 | -0.3880 | No |
| 208 | RFC1 | RFC1 Entrez, Source | replication factor C (activator 1) 1, 145kDa | 10535 | -0.046 | -0.3867 | No |
| 209 | USP11 | USP11 Entrez, Source | ubiquitin specific peptidase 11 | 10587 | -0.047 | -0.3883 | No |
| 210 | MRPL49 | MRPL49 Entrez, Source | mitochondrial ribosomal protein L49 | 10593 | -0.047 | -0.3864 | No |
| 211 | CDC25B | CDC25B Entrez, Source | cell division cycle 25B | 10619 | -0.047 | -0.3860 | No |
| 212 | DR1 | DR1 Entrez, Source | down-regulator of transcription 1, TBP-binding (negative cofactor 2) | 10633 | -0.048 | -0.3847 | No |
| 213 | STXBP3 | STXBP3 Entrez, Source | syntaxin binding protein 3 | 10705 | -0.050 | -0.3877 | No |
| 214 | KIF5B | KIF5B Entrez, Source | kinesin family member 5B | 10742 | -0.050 | -0.3880 | No |
| 215 | CBR1 | CBR1 Entrez, Source | carbonyl reductase 1 | 10804 | -0.052 | -0.3901 | No |
| 216 | PLCD1 | PLCD1 Entrez, Source | phospholipase C, delta 1 | 10835 | -0.052 | -0.3899 | No |
| 217 | CKAP5 | CKAP5 Entrez, Source | cytoskeleton associated protein 5 | 10855 | -0.053 | -0.3888 | No |
| 218 | TGFBI | TGFBI Entrez, Source | transforming growth factor, beta-induced, 68kDa | 10868 | -0.053 | -0.3871 | No |
| 219 | ABCD3 | ABCD3 Entrez, Source | ATP-binding cassette, sub-family D (ALD), member 3 | 10872 | -0.053 | -0.3847 | No |
| 220 | EPS15 | EPS15 Entrez, Source | epidermal growth factor receptor pathway substrate 15 | 11024 | -0.056 | -0.3935 | No |
| 221 | FNTA | FNTA Entrez, Source | farnesyltransferase, CAAX box, alpha | 11067 | -0.057 | -0.3939 | No |
| 222 | RELA | RELA Entrez, Source | v-rel reticuloendotheliosis viral oncogene homolog A, nuclear factor of kappa light polypeptide gene enhancer in B-cells 3, p65 (avian) | 11138 | -0.059 | -0.3964 | No |
| 223 | HIF1A | HIF1A Entrez, Source | hypoxia-inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) | 11170 | -0.060 | -0.3959 | No |
| 224 | CLTA | CLTA Entrez, Source | clathrin, light chain (Lca) | 11175 | -0.060 | -0.3932 | No |
| 225 | MAP2K3 | MAP2K3 Entrez, Source | mitogen-activated protein kinase kinase 3 | 11186 | -0.060 | -0.3911 | No |
| 226 | GTF3C2 | GTF3C2 Entrez, Source | general transcription factor IIIC, polypeptide 2, beta 110kDa | 11404 | -0.065 | -0.4045 | No |
| 227 | NEU1 | NEU1 Entrez, Source | sialidase 1 (lysosomal sialidase) | 11548 | -0.070 | -0.4120 | No |
| 228 | GSK3B | GSK3B Entrez, Source | glycogen synthase kinase 3 beta | 11670 | -0.074 | -0.4176 | No |
| 229 | CIRBP | CIRBP Entrez, Source | cold inducible RNA binding protein | 11768 | -0.078 | -0.4212 | Yes |
| 230 | PPP4R1 | PPP4R1 Entrez, Source | protein phosphatase 4, regulatory subunit 1 | 11788 | -0.078 | -0.4188 | Yes |
| 231 | APP | APP Entrez, Source | amyloid beta (A4) precursor protein (peptidase nexin-II, Alzheimer disease) | 11831 | -0.081 | -0.4181 | Yes |
| 232 | H2AFX | H2AFX Entrez, Source | H2A histone family, member X | 11841 | -0.081 | -0.4148 | Yes |
| 233 | CD99 | CD99 Entrez, Source | CD99 molecule | 11871 | -0.082 | -0.4131 | Yes |
| 234 | CXADR | CXADR Entrez, Source | coxsackie virus and adenovirus receptor | 11909 | -0.084 | -0.4118 | Yes |
| 235 | CDC37 | CDC37 Entrez, Source | CDC37 cell division cycle 37 homolog (S. cerevisiae) | 11983 | -0.086 | -0.4132 | Yes |
| 236 | STK3 | STK3 Entrez, Source | serine/threonine kinase 3 (STE20 homolog, yeast) | 11999 | -0.087 | -0.4101 | Yes |
| 237 | CBFB | CBFB Entrez, Source | core-binding factor, beta subunit | 12204 | -0.097 | -0.4209 | Yes |
| 238 | DYNLL1 | DYNLL1 Entrez, Source | dynein, light chain, LC8-type 1 | 12221 | -0.098 | -0.4173 | Yes |
| 239 | QSCN6 | QSCN6 Entrez, Source | quiescin Q6 | 12235 | -0.099 | -0.4135 | Yes |
| 240 | TPMT | TPMT Entrez, Source | thiopurine S-methyltransferase | 12268 | -0.101 | -0.4110 | Yes |
| 241 | IL1R2 | IL1R2 Entrez, Source | interleukin 1 receptor, type II | 12286 | -0.103 | -0.4073 | Yes |
| 242 | SFRS9 | SFRS9 Entrez, Source | splicing factor, arginine/serine-rich 9 | 12292 | -0.103 | -0.4026 | Yes |
| 243 | TJP2 | TJP2 Entrez, Source | tight junction protein 2 (zona occludens 2) | 12313 | -0.104 | -0.3991 | Yes |
| 244 | SRD5A1 | SRD5A1 Entrez, Source | steroid-5-alpha-reductase, alpha polypeptide 1 (3-oxo-5 alpha-steroid delta 4-dehydrogenase alpha 1) | 12386 | -0.110 | -0.3992 | Yes |
| 245 | TFPI2 | TFPI2 Entrez, Source | tissue factor pathway inhibitor 2 | 12427 | -0.112 | -0.3967 | Yes |
| 246 | MYH10 | MYH10 Entrez, Source | myosin, heavy chain 10, non-muscle | 12486 | -0.117 | -0.3955 | Yes |
| 247 | EMP1 | EMP1 Entrez, Source | epithelial membrane protein 1 | 12519 | -0.119 | -0.3921 | Yes |
| 248 | CYR61 | CYR61 Entrez, Source | cysteine-rich, angiogenic inducer, 61 | 12557 | -0.122 | -0.3890 | Yes |
| 249 | CDK5 | CDK5 Entrez, Source | cyclin-dependent kinase 5 | 12579 | -0.124 | -0.3845 | Yes |
| 250 | PTPRF | PTPRF Entrez, Source | protein tyrosine phosphatase, receptor type, F | 12684 | -0.133 | -0.3859 | Yes |
| 251 | ITGA6 | ITGA6 Entrez, Source | integrin, alpha 6 | 12716 | -0.137 | -0.3816 | Yes |
| 252 | TSC22D1 | TSC22D1 Entrez, Source | TSC22 domain family, member 1 | 12825 | -0.152 | -0.3825 | Yes |
| 253 | PTPRZ1 | PTPRZ1 Entrez, Source | protein tyrosine phosphatase, receptor-type, Z polypeptide 1 | 12829 | -0.152 | -0.3752 | Yes |
| 254 | ABCC1 | ABCC1 Entrez, Source | ATP-binding cassette, sub-family C (CFTR/MRP), member 1 | 12872 | -0.160 | -0.3706 | Yes |
| 255 | TAX1BP3 | TAX1BP3 Entrez, Source | Tax1 (human T-cell leukemia virus type I) binding protein 3 | 12900 | -0.165 | -0.3646 | Yes |
| 256 | CST6 | CST6 Entrez, Source | cystatin E/M | 12920 | -0.170 | -0.3577 | Yes |
| 257 | TRIP10 | TRIP10 Entrez, Source | thyroid hormone receptor interactor 10 | 12922 | -0.170 | -0.3494 | Yes |
| 258 | HBEGF | HBEGF Entrez, Source | heparin-binding EGF-like growth factor | 12974 | -0.182 | -0.3444 | Yes |
| 259 | VGLL4 | VGLL4 Entrez, Source | vestigial like 4 (Drosophila) | 12975 | -0.182 | -0.3355 | Yes |
| 260 | ALCAM | ALCAM Entrez, Source | activated leukocyte cell adhesion molecule | 12991 | -0.186 | -0.3275 | Yes |
| 261 | SCRN1 | SCRN1 Entrez, Source | secernin 1 | 12993 | -0.186 | -0.3185 | Yes |
| 262 | UGCG | UGCG Entrez, Source | UDP-glucose ceramide glucosyltransferase | 13105 | -0.234 | -0.3155 | Yes |
| 263 | JAG1 | JAG1 Entrez, Source | jagged 1 (Alagille syndrome) | 13149 | -0.256 | -0.3063 | Yes |
| 264 | CRYAB | CRYAB Entrez, Source | crystallin, alpha B | 13206 | -0.298 | -0.2960 | Yes |
| 265 | SLC2A1 | SLC2A1 Entrez, Source | solute carrier family 2 (facilitated glucose transporter), member 1 | 13210 | -0.303 | -0.2814 | Yes |
| 266 | PAM | PAM Entrez, Source | peptidylglycine alpha-amidating monooxygenase | 13213 | -0.306 | -0.2666 | Yes |
| 267 | ID3 | ID3 Entrez, Source | inhibitor of DNA binding 3, dominant negative helix-loop-helix protein | 13220 | -0.314 | -0.2516 | Yes |
| 268 | CITED2 | CITED2 Entrez, Source | Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 | 13226 | -0.322 | -0.2363 | Yes |
| 269 | GNAI1 | GNAI1 Entrez, Source | guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 | 13239 | -0.335 | -0.2208 | Yes |
| 270 | MME | MME Entrez, Source | membrane metallo-endopeptidase (neutral endopeptidase, enkephalinase) | 13245 | -0.347 | -0.2041 | Yes |
| 271 | ADM | ADM Entrez, Source | adrenomedullin | 13274 | -0.393 | -0.1870 | Yes |
| 272 | TPBG | TPBG Entrez, Source | trophoblast glycoprotein | 13288 | -0.427 | -0.1671 | Yes |
| 273 | SLC31A2 | SLC31A2 Entrez, Source | solute carrier family 31 (copper transporters), member 2 | 13300 | -0.468 | -0.1451 | Yes |
| 274 | LTBP1 | LTBP1 Entrez, Source | latent transforming growth factor beta binding protein 1 | 13307 | -0.479 | -0.1221 | Yes |
| 275 | ANXA1 | ANXA1 Entrez, Source | annexin A1 | 13312 | -0.498 | -0.0980 | Yes |
| 276 | CTGF | CTGF Entrez, Source | connective tissue growth factor | 13313 | -0.503 | -0.0734 | Yes |
| 277 | F3 | F3 Entrez, Source | coagulation factor III (thromboplastin, tissue factor) | 13315 | -0.511 | -0.0484 | Yes |
| 278 | KIAA0101 | KIAA0101 Entrez, Source | KIAA0101 | 13316 | -0.512 | -0.0233 | Yes |
| 279 | PFKP | PFKP Entrez, Source | phosphofructokinase, platelet | 13318 | -0.516 | 0.0018 | Yes |