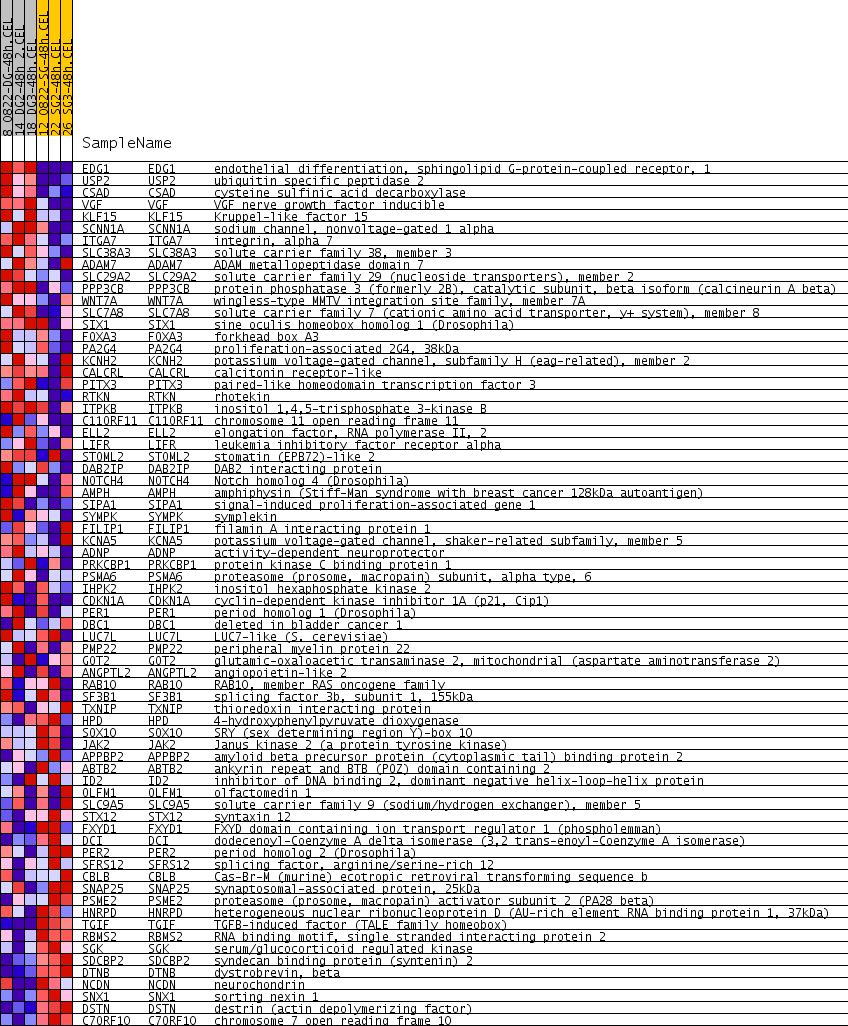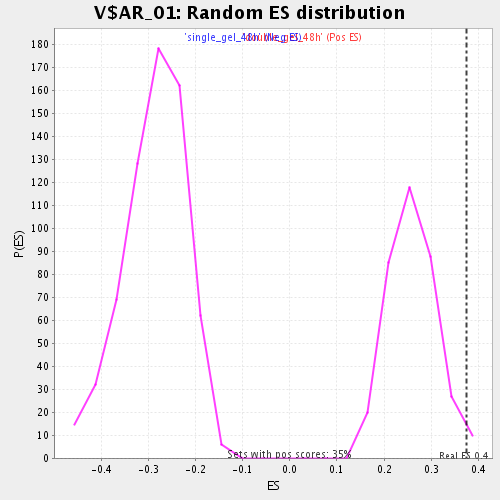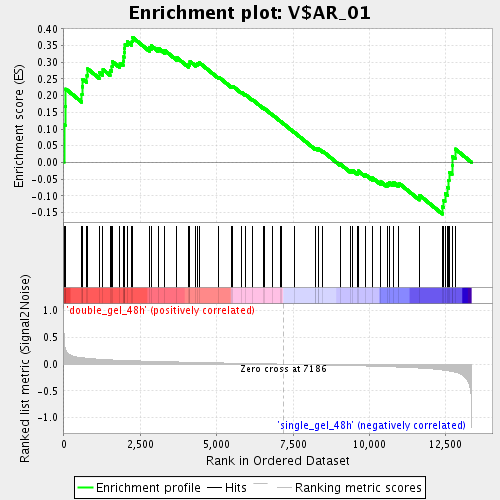
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_48h_versus_single_gel_48h.class.cls #double_gel_48h_versus_single_gel_48h_repos |
| Phenotype | class.cls#double_gel_48h_versus_single_gel_48h_repos |
| Upregulated in class | double_gel_48h |
| GeneSet | V$AR_01 |
| Enrichment Score (ES) | 0.37504724 |
| Normalized Enrichment Score (NES) | 1.4510049 |
| Nominal p-value | 0.01724138 |
| FDR q-value | 0.17630257 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | EDG1 | EDG1 Entrez, Source | endothelial differentiation, sphingolipid G-protein-coupled receptor, 1 | 3 | 0.566 | 0.1128 | Yes |
| 2 | USP2 | USP2 Entrez, Source | ubiquitin specific peptidase 2 | 42 | 0.293 | 0.1686 | Yes |
| 3 | CSAD | CSAD Entrez, Source | cysteine sulfinic acid decarboxylase | 62 | 0.265 | 0.2201 | Yes |
| 4 | VGF | VGF Entrez, Source | VGF nerve growth factor inducible | 587 | 0.119 | 0.2044 | Yes |
| 5 | KLF15 | KLF15 Entrez, Source | Kruppel-like factor 15 | 600 | 0.118 | 0.2270 | Yes |
| 6 | SCNN1A | SCNN1A Entrez, Source | sodium channel, nonvoltage-gated 1 alpha | 616 | 0.117 | 0.2492 | Yes |
| 7 | ITGA7 | ITGA7 Entrez, Source | integrin, alpha 7 | 752 | 0.109 | 0.2608 | Yes |
| 8 | SLC38A3 | SLC38A3 Entrez, Source | solute carrier family 38, member 3 | 779 | 0.108 | 0.2803 | Yes |
| 9 | ADAM7 | ADAM7 Entrez, Source | ADAM metallopeptidase domain 7 | 1156 | 0.091 | 0.2702 | Yes |
| 10 | SLC29A2 | SLC29A2 Entrez, Source | solute carrier family 29 (nucleoside transporters), member 2 | 1277 | 0.087 | 0.2785 | Yes |
| 11 | PPP3CB | PPP3CB Entrez, Source | protein phosphatase 3 (formerly 2B), catalytic subunit, beta isoform (calcineurin A beta) | 1512 | 0.079 | 0.2766 | Yes |
| 12 | WNT7A | WNT7A Entrez, Source | wingless-type MMTV integration site family, member 7A | 1571 | 0.078 | 0.2878 | Yes |
| 13 | SLC7A8 | SLC7A8 Entrez, Source | solute carrier family 7 (cationic amino acid transporter, y+ system), member 8 | 1584 | 0.077 | 0.3023 | Yes |
| 14 | SIX1 | SIX1 Entrez, Source | sine oculis homeobox homolog 1 (Drosophila) | 1834 | 0.071 | 0.2978 | Yes |
| 15 | FOXA3 | FOXA3 Entrez, Source | forkhead box A3 | 1938 | 0.069 | 0.3039 | Yes |
| 16 | PA2G4 | PA2G4 Entrez, Source | proliferation-associated 2G4, 38kDa | 1958 | 0.069 | 0.3163 | Yes |
| 17 | KCNH2 | KCNH2 Entrez, Source | potassium voltage-gated channel, subfamily H (eag-related), member 2 | 1971 | 0.069 | 0.3291 | Yes |
| 18 | CALCRL | CALCRL Entrez, Source | calcitonin receptor-like | 1986 | 0.069 | 0.3418 | Yes |
| 19 | PITX3 | PITX3 Entrez, Source | paired-like homeodomain transcription factor 3 | 1998 | 0.068 | 0.3546 | Yes |
| 20 | RTKN | RTKN Entrez, Source | rhotekin | 2071 | 0.066 | 0.3624 | Yes |
| 21 | ITPKB | ITPKB Entrez, Source | inositol 1,4,5-trisphosphate 3-kinase B | 2227 | 0.064 | 0.3634 | Yes |
| 22 | C11ORF11 | C11ORF11 Entrez, Source | chromosome 11 open reading frame 11 | 2241 | 0.063 | 0.3750 | Yes |
| 23 | ELL2 | ELL2 Entrez, Source | elongation factor, RNA polymerase II, 2 | 2797 | 0.053 | 0.3439 | No |
| 24 | LIFR | LIFR Entrez, Source | leukemia inhibitory factor receptor alpha | 2859 | 0.052 | 0.3497 | No |
| 25 | STOML2 | STOML2 Entrez, Source | stomatin (EPB72)-like 2 | 3084 | 0.049 | 0.3426 | No |
| 26 | DAB2IP | DAB2IP Entrez, Source | DAB2 interacting protein | 3298 | 0.046 | 0.3357 | No |
| 27 | NOTCH4 | NOTCH4 Entrez, Source | Notch homolog 4 (Drosophila) | 3694 | 0.041 | 0.3141 | No |
| 28 | AMPH | AMPH Entrez, Source | amphiphysin (Stiff-Man syndrome with breast cancer 128kDa autoantigen) | 4075 | 0.035 | 0.2925 | No |
| 29 | SIPA1 | SIPA1 Entrez, Source | signal-induced proliferation-associated gene 1 | 4121 | 0.035 | 0.2961 | No |
| 30 | SYMPK | SYMPK Entrez, Source | symplekin | 4123 | 0.035 | 0.3029 | No |
| 31 | FILIP1 | FILIP1 Entrez, Source | filamin A interacting protein 1 | 4318 | 0.032 | 0.2948 | No |
| 32 | KCNA5 | KCNA5 Entrez, Source | potassium voltage-gated channel, shaker-related subfamily, member 5 | 4364 | 0.032 | 0.2977 | No |
| 33 | ADNP | ADNP Entrez, Source | activity-dependent neuroprotector | 4421 | 0.031 | 0.2997 | No |
| 34 | PRKCBP1 | PRKCBP1 Entrez, Source | protein kinase C binding protein 1 | 5072 | 0.023 | 0.2553 | No |
| 35 | PSMA6 | PSMA6 Entrez, Source | proteasome (prosome, macropain) subunit, alpha type, 6 | 5475 | 0.019 | 0.2287 | No |
| 36 | IHPK2 | IHPK2 Entrez, Source | inositol hexaphosphate kinase 2 | 5529 | 0.018 | 0.2283 | No |
| 37 | CDKN1A | CDKN1A Entrez, Source | cyclin-dependent kinase inhibitor 1A (p21, Cip1) | 5802 | 0.015 | 0.2108 | No |
| 38 | PER1 | PER1 Entrez, Source | period homolog 1 (Drosophila) | 5944 | 0.013 | 0.2029 | No |
| 39 | DBC1 | DBC1 Entrez, Source | deleted in bladder cancer 1 | 6173 | 0.011 | 0.1879 | No |
| 40 | LUC7L | LUC7L Entrez, Source | LUC7-like (S. cerevisiae) | 6518 | 0.007 | 0.1635 | No |
| 41 | PMP22 | PMP22 Entrez, Source | peripheral myelin protein 22 | 6574 | 0.007 | 0.1607 | No |
| 42 | GOT2 | GOT2 Entrez, Source | glutamic-oxaloacetic transaminase 2, mitochondrial (aspartate aminotransferase 2) | 6812 | 0.004 | 0.1436 | No |
| 43 | ANGPTL2 | ANGPTL2 Entrez, Source | angiopoietin-like 2 | 7080 | 0.001 | 0.1238 | No |
| 44 | RAB10 | RAB10 Entrez, Source | RAB10, member RAS oncogene family | 7106 | 0.001 | 0.1220 | No |
| 45 | SF3B1 | SF3B1 Entrez, Source | splicing factor 3b, subunit 1, 155kDa | 7535 | -0.004 | 0.0906 | No |
| 46 | TXNIP | TXNIP Entrez, Source | thioredoxin interacting protein | 8238 | -0.012 | 0.0402 | No |
| 47 | HPD | HPD Entrez, Source | 4-hydroxyphenylpyruvate dioxygenase | 8248 | -0.012 | 0.0420 | No |
| 48 | SOX10 | SOX10 Entrez, Source | SRY (sex determining region Y)-box 10 | 8326 | -0.013 | 0.0388 | No |
| 49 | JAK2 | JAK2 Entrez, Source | Janus kinase 2 (a protein tyrosine kinase) | 8330 | -0.013 | 0.0413 | No |
| 50 | APPBP2 | APPBP2 Entrez, Source | amyloid beta precursor protein (cytoplasmic tail) binding protein 2 | 8474 | -0.015 | 0.0335 | No |
| 51 | ABTB2 | ABTB2 Entrez, Source | ankyrin repeat and BTB (POZ) domain containing 2 | 9037 | -0.022 | -0.0044 | No |
| 52 | ID2 | ID2 Entrez, Source | inhibitor of DNA binding 2, dominant negative helix-loop-helix protein | 9384 | -0.027 | -0.0250 | No |
| 53 | OLFM1 | OLFM1 Entrez, Source | olfactomedin 1 | 9453 | -0.028 | -0.0245 | No |
| 54 | SLC9A5 | SLC9A5 Entrez, Source | solute carrier family 9 (sodium/hydrogen exchanger), member 5 | 9614 | -0.031 | -0.0304 | No |
| 55 | STX12 | STX12 Entrez, Source | syntaxin 12 | 9628 | -0.031 | -0.0252 | No |
| 56 | FXYD1 | FXYD1 Entrez, Source | FXYD domain containing ion transport regulator 1 (phospholemman) | 9863 | -0.034 | -0.0360 | No |
| 57 | DCI | DCI Entrez, Source | dodecenoyl-Coenzyme A delta isomerase (3,2 trans-enoyl-Coenzyme A isomerase) | 10098 | -0.038 | -0.0460 | No |
| 58 | PER2 | PER2 Entrez, Source | period homolog 2 (Drosophila) | 10374 | -0.043 | -0.0581 | No |
| 59 | SFRS12 | SFRS12 Entrez, Source | splicing factor, arginine/serine-rich 12 | 10579 | -0.047 | -0.0642 | No |
| 60 | CBLB | CBLB Entrez, Source | Cas-Br-M (murine) ecotropic retroviral transforming sequence b | 10642 | -0.048 | -0.0593 | No |
| 61 | SNAP25 | SNAP25 Entrez, Source | synaptosomal-associated protein, 25kDa | 10791 | -0.051 | -0.0602 | No |
| 62 | PSME2 | PSME2 Entrez, Source | proteasome (prosome, macropain) activator subunit 2 (PA28 beta) | 10964 | -0.055 | -0.0622 | No |
| 63 | HNRPD | HNRPD Entrez, Source | heterogeneous nuclear ribonucleoprotein D (AU-rich element RNA binding protein 1, 37kDa) | 11634 | -0.073 | -0.0979 | No |
| 64 | TGIF | TGIF Entrez, Source | TGFB-induced factor (TALE family homeobox) | 12388 | -0.110 | -0.1327 | No |
| 65 | RBMS2 | RBMS2 Entrez, Source | RNA binding motif, single stranded interacting protein 2 | 12431 | -0.113 | -0.1133 | No |
| 66 | SGK | SGK Entrez, Source | serum/glucocorticoid regulated kinase | 12473 | -0.116 | -0.0933 | No |
| 67 | SDCBP2 | SDCBP2 Entrez, Source | syndecan binding protein (syntenin) 2 | 12560 | -0.122 | -0.0754 | No |
| 68 | DTNB | DTNB Entrez, Source | dystrobrevin, beta | 12592 | -0.125 | -0.0528 | No |
| 69 | NCDN | NCDN Entrez, Source | neurochondrin | 12607 | -0.126 | -0.0288 | No |
| 70 | SNX1 | SNX1 Entrez, Source | sorting nexin 1 | 12703 | -0.136 | -0.0089 | No |
| 71 | DSTN | DSTN Entrez, Source | destrin (actin depolymerizing factor) | 12710 | -0.136 | 0.0179 | No |
| 72 | C7ORF10 | C7ORF10 Entrez, Source | chromosome 7 open reading frame 10 | 12806 | -0.149 | 0.0404 | No |