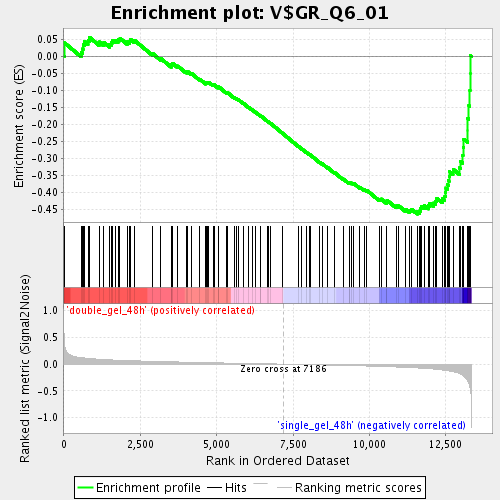
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_48h_versus_single_gel_48h.class.cls #double_gel_48h_versus_single_gel_48h_repos |
| Phenotype | class.cls#double_gel_48h_versus_single_gel_48h_repos |
| Upregulated in class | single_gel_48h |
| GeneSet | V$GR_Q6_01 |
| Enrichment Score (ES) | -0.46489632 |
| Normalized Enrichment Score (NES) | -1.7015887 |
| Nominal p-value | 0.0014880953 |
| FDR q-value | 0.06627474 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | WNT4 | WNT4 Entrez, Source | wingless-type MMTV integration site family, member 4 | 16 | 0.365 | 0.0405 | No |
| 2 | LGI1 | LGI1 Entrez, Source | leucine-rich, glioma inactivated 1 | 570 | 0.120 | 0.0124 | No |
| 3 | SCNN1A | SCNN1A Entrez, Source | sodium channel, nonvoltage-gated 1 alpha | 616 | 0.117 | 0.0224 | No |
| 4 | C6ORF66 | C6ORF66 Entrez, Source | chromosome 6 open reading frame 66 | 630 | 0.116 | 0.0347 | No |
| 5 | PCDH8 | PCDH8 Entrez, Source | protocadherin 8 | 673 | 0.114 | 0.0445 | No |
| 6 | SLC25A10 | SLC25A10 Entrez, Source | solute carrier family 25 (mitochondrial carrier; dicarboxylate transporter), member 10 | 790 | 0.107 | 0.0480 | No |
| 7 | EPHA7 | EPHA7 Entrez, Source | EPH receptor A7 | 833 | 0.105 | 0.0568 | No |
| 8 | SULT2A1 | SULT2A1 Entrez, Source | sulfotransferase family, cytosolic, 2A, dehydroepiandrosterone (DHEA)-preferring, member 1 | 1153 | 0.091 | 0.0431 | No |
| 9 | LTBP3 | LTBP3 Entrez, Source | latent transforming growth factor beta binding protein 3 | 1306 | 0.086 | 0.0415 | No |
| 10 | ME1 | ME1 Entrez, Source | malic enzyme 1, NADP(+)-dependent, cytosolic | 1493 | 0.080 | 0.0365 | No |
| 11 | PPP1R10 | PPP1R10 Entrez, Source | protein phosphatase 1, regulatory subunit 10 | 1553 | 0.078 | 0.0410 | No |
| 12 | OPN1SW | OPN1SW Entrez, Source | opsin 1 (cone pigments), short-wave-sensitive (color blindness, tritan) | 1576 | 0.078 | 0.0482 | No |
| 13 | HOXA2 | HOXA2 Entrez, Source | homeobox A2 | 1685 | 0.075 | 0.0485 | No |
| 14 | DND1 | DND1 Entrez, Source | dead end homolog 1 (zebrafish) | 1773 | 0.073 | 0.0502 | No |
| 15 | SIX1 | SIX1 Entrez, Source | sine oculis homeobox homolog 1 (Drosophila) | 1834 | 0.071 | 0.0539 | No |
| 16 | RTKN | RTKN Entrez, Source | rhotekin | 2071 | 0.066 | 0.0436 | No |
| 17 | AKAP5 | AKAP5 Entrez, Source | A kinase (PRKA) anchor protein 5 | 2147 | 0.065 | 0.0454 | No |
| 18 | CDH2 | CDH2 Entrez, Source | cadherin 2, type 1, N-cadherin (neuronal) | 2163 | 0.065 | 0.0516 | No |
| 19 | TNNT3 | TNNT3 Entrez, Source | troponin T type 3 (skeletal, fast) | 2298 | 0.062 | 0.0486 | No |
| 20 | CSNK1G1 | CSNK1G1 Entrez, Source | casein kinase 1, gamma 1 | 2899 | 0.052 | 0.0091 | No |
| 21 | GRP | GRP Entrez, Source | gastrin-releasing peptide | 3174 | 0.048 | -0.0061 | No |
| 22 | TMEM24 | TMEM24 Entrez, Source | transmembrane protein 24 | 3509 | 0.043 | -0.0265 | No |
| 23 | PRG1 | PRG1 Entrez, Source | proteoglycan 1, secretory granule | 3518 | 0.043 | -0.0222 | No |
| 24 | SMAD6 | SMAD6 Entrez, Source | SMAD, mothers against DPP homolog 6 (Drosophila) | 3551 | 0.042 | -0.0198 | No |
| 25 | BAT4 | BAT4 Entrez, Source | HLA-B associated transcript 4 | 3719 | 0.040 | -0.0278 | No |
| 26 | MPZ | MPZ Entrez, Source | myelin protein zero (Charcot-Marie-Tooth neuropathy 1B) | 3998 | 0.036 | -0.0447 | No |
| 27 | MRPS18B | MRPS18B Entrez, Source | mitochondrial ribosomal protein S18B | 4053 | 0.036 | -0.0447 | No |
| 28 | NCOA3 | NCOA3 Entrez, Source | nuclear receptor coactivator 3 | 4160 | 0.034 | -0.0488 | No |
| 29 | SLC18A3 | SLC18A3 Entrez, Source | solute carrier family 18 (vesicular acetylcholine), member 3 | 4440 | 0.031 | -0.0664 | No |
| 30 | ANK3 | ANK3 Entrez, Source | ankyrin 3, node of Ranvier (ankyrin G) | 4640 | 0.028 | -0.0782 | No |
| 31 | LRRN6A | LRRN6A Entrez, Source | leucine rich repeat neuronal 6A | 4672 | 0.028 | -0.0773 | No |
| 32 | KCNK10 | KCNK10 Entrez, Source | potassium channel, subfamily K, member 10 | 4698 | 0.028 | -0.0761 | No |
| 33 | EIF4G2 | EIF4G2 Entrez, Source | eukaryotic translation initiation factor 4 gamma, 2 | 4729 | 0.027 | -0.0753 | No |
| 34 | PROK2 | PROK2 Entrez, Source | prokineticin 2 | 4883 | 0.025 | -0.0839 | No |
| 35 | NOG | NOG Entrez, Source | noggin | 4913 | 0.025 | -0.0833 | No |
| 36 | GMPR2 | GMPR2 Entrez, Source | guanosine monophosphate reductase 2 | 5045 | 0.023 | -0.0905 | No |
| 37 | AKAP7 | AKAP7 Entrez, Source | A kinase (PRKA) anchor protein 7 | 5056 | 0.023 | -0.0886 | No |
| 38 | SYNCRIP | SYNCRIP Entrez, Source | synaptotagmin binding, cytoplasmic RNA interacting protein | 5319 | 0.020 | -0.1061 | No |
| 39 | MASP1 | MASP1 Entrez, Source | mannan-binding lectin serine peptidase 1 (C4/C2 activating component of Ra-reactive factor) | 5355 | 0.020 | -0.1065 | No |
| 40 | APBA1 | APBA1 Entrez, Source | amyloid beta (A4) precursor protein-binding, family A, member 1 (X11) | 5578 | 0.017 | -0.1213 | No |
| 41 | OTX2 | OTX2 Entrez, Source | orthodenticle homolog 2 (Drosophila) | 5633 | 0.017 | -0.1235 | No |
| 42 | EYA2 | EYA2 Entrez, Source | eyes absent homolog 2 (Drosophila) | 5703 | 0.016 | -0.1268 | No |
| 43 | ZP3 | ZP3 Entrez, Source | zona pellucida glycoprotein 3 (sperm receptor) | 5866 | 0.014 | -0.1375 | No |
| 44 | EIF2B4 | EIF2B4 Entrez, Source | eukaryotic translation initiation factor 2B, subunit 4 delta, 67kDa | 6048 | 0.012 | -0.1497 | No |
| 45 | PPM1B | PPM1B Entrez, Source | protein phosphatase 1B (formerly 2C), magnesium-dependent, beta isoform | 6165 | 0.011 | -0.1572 | No |
| 46 | CPA3 | CPA3 Entrez, Source | carboxypeptidase A3 (mast cell) | 6266 | 0.010 | -0.1636 | No |
| 47 | MYH4 | MYH4 Entrez, Source | myosin, heavy chain 4, skeletal muscle | 6427 | 0.008 | -0.1748 | No |
| 48 | CREBBP | CREBBP Entrez, Source | CREB binding protein (Rubinstein-Taybi syndrome) | 6449 | 0.008 | -0.1754 | No |
| 49 | SV2B | SV2B Entrez, Source | synaptic vesicle glycoprotein 2B | 6659 | 0.006 | -0.1906 | No |
| 50 | SLC2A2 | SLC2A2 Entrez, Source | solute carrier family 2 (facilitated glucose transporter), member 2 | 6701 | 0.005 | -0.1930 | No |
| 51 | FGD4 | FGD4 Entrez, Source | FYVE, RhoGEF and PH domain containing 4 | 6756 | 0.005 | -0.1966 | No |
| 52 | EFNA1 | EFNA1 Entrez, Source | ephrin-A1 | 7146 | 0.000 | -0.2259 | No |
| 53 | SYT6 | SYT6 Entrez, Source | synaptotagmin VI | 7678 | -0.006 | -0.2654 | No |
| 54 | RBX1 | RBX1 Entrez, Source | ring-box 1 | 7789 | -0.007 | -0.2729 | No |
| 55 | PDAP1 | PDAP1 Entrez, Source | PDGFA associated protein 1 | 7929 | -0.009 | -0.2824 | No |
| 56 | AQP4 | AQP4 Entrez, Source | aquaporin 4 | 8023 | -0.010 | -0.2884 | No |
| 57 | PAX3 | PAX3 Entrez, Source | paired box gene 3 (Waardenburg syndrome 1) | 8060 | -0.010 | -0.2899 | No |
| 58 | FEN1 | FEN1 Entrez, Source | flap structure-specific endonuclease 1 | 8375 | -0.014 | -0.3121 | No |
| 59 | ST13 | ST13 Entrez, Source | suppression of tumorigenicity 13 (colon carcinoma) (Hsp70 interacting protein) | 8448 | -0.015 | -0.3158 | No |
| 60 | EP300 | EP300 Entrez, Source | E1A binding protein p300 | 8639 | -0.017 | -0.3282 | No |
| 61 | QKI | QKI Entrez, Source | quaking homolog, KH domain RNA binding (mouse) | 8852 | -0.020 | -0.3419 | No |
| 62 | PDYN | PDYN Entrez, Source | prodynorphin | 9146 | -0.024 | -0.3614 | No |
| 63 | FANK1 | FANK1 Entrez, Source | fibronectin type III and ankyrin repeat domains 1 | 9339 | -0.026 | -0.3728 | No |
| 64 | ABCG8 | ABCG8 Entrez, Source | ATP-binding cassette, sub-family G (WHITE), member 8 (sterolin 2) | 9348 | -0.027 | -0.3704 | No |
| 65 | TNMD | TNMD Entrez, Source | tenomodulin | 9425 | -0.028 | -0.3730 | No |
| 66 | OTX1 | OTX1 Entrez, Source | orthodenticle homolog 1 (Drosophila) | 9487 | -0.029 | -0.3743 | No |
| 67 | NRP2 | NRP2 Entrez, Source | neuropilin 2 | 9682 | -0.032 | -0.3853 | No |
| 68 | NEDD8 | NEDD8 Entrez, Source | neural precursor cell expressed, developmentally down-regulated 8 | 9830 | -0.034 | -0.3926 | No |
| 69 | SLC38A2 | SLC38A2 Entrez, Source | solute carrier family 38, member 2 | 9913 | -0.035 | -0.3948 | No |
| 70 | ITPR3 | ITPR3 Entrez, Source | inositol 1,4,5-triphosphate receptor, type 3 | 10318 | -0.042 | -0.4205 | No |
| 71 | ZNF24 | ZNF24 Entrez, Source | zinc finger protein 24 | 10380 | -0.043 | -0.4202 | No |
| 72 | CXCR4 | CXCR4 Entrez, Source | chemokine (C-X-C motif) receptor 4 | 10564 | -0.046 | -0.4288 | No |
| 73 | PSEN1 | PSEN1 Entrez, Source | presenilin 1 (Alzheimer disease 3) | 10571 | -0.046 | -0.4239 | No |
| 74 | KLHL5 | KLHL5 Entrez, Source | kelch-like 5 (Drosophila) | 10873 | -0.053 | -0.4406 | No |
| 75 | PACSIN3 | PACSIN3 Entrez, Source | protein kinase C and casein kinase substrate in neurons 3 | 10954 | -0.055 | -0.4404 | No |
| 76 | TCN2 | TCN2 Entrez, Source | transcobalamin II; macrocytic anemia | 11172 | -0.060 | -0.4500 | No |
| 77 | SNX17 | SNX17 Entrez, Source | sorting nexin 17 | 11315 | -0.063 | -0.4535 | No |
| 78 | RND1 | RND1 Entrez, Source | Rho family GTPase 1 | 11382 | -0.065 | -0.4511 | No |
| 79 | NPEPPS | NPEPPS Entrez, Source | aminopeptidase puromycin sensitive | 11565 | -0.071 | -0.4568 | Yes |
| 80 | CDKN1B | CDKN1B Entrez, Source | cyclin-dependent kinase inhibitor 1B (p27, Kip1) | 11651 | -0.074 | -0.4548 | Yes |
| 81 | KHDRBS1 | KHDRBS1 Entrez, Source | KH domain containing, RNA binding, signal transduction associated 1 | 11672 | -0.074 | -0.4478 | Yes |
| 82 | SDC1 | SDC1 Entrez, Source | syndecan 1 | 11699 | -0.075 | -0.4412 | Yes |
| 83 | MMP14 | MMP14 Entrez, Source | matrix metallopeptidase 14 (membrane-inserted) | 11798 | -0.079 | -0.4396 | Yes |
| 84 | LASP1 | LASP1 Entrez, Source | LIM and SH3 protein 1 | 11935 | -0.084 | -0.4402 | Yes |
| 85 | FOXA1 | FOXA1 Entrez, Source | forkhead box A1 | 11968 | -0.086 | -0.4329 | Yes |
| 86 | ARG2 | ARG2 Entrez, Source | arginase, type II | 12097 | -0.091 | -0.4321 | Yes |
| 87 | CAMLG | CAMLG Entrez, Source | calcium modulating ligand | 12165 | -0.095 | -0.4264 | Yes |
| 88 | SF3B4 | SF3B4 Entrez, Source | splicing factor 3b, subunit 4, 49kDa | 12200 | -0.097 | -0.4179 | Yes |
| 89 | TGIF | TGIF Entrez, Source | TGFB-induced factor (TALE family homeobox) | 12388 | -0.110 | -0.4194 | Yes |
| 90 | JMJD1C | JMJD1C Entrez, Source | jumonji domain containing 1C | 12465 | -0.115 | -0.4120 | Yes |
| 91 | MAP1B | MAP1B Entrez, Source | microtubule-associated protein 1B | 12487 | -0.117 | -0.4003 | Yes |
| 92 | DHX40 | DHX40 Entrez, Source | DEAH (Asp-Glu-Ala-His) box polypeptide 40 | 12501 | -0.117 | -0.3879 | Yes |
| 93 | MAGED1 | MAGED1 Entrez, Source | melanoma antigen family D, 1 | 12552 | -0.121 | -0.3778 | Yes |
| 94 | PLSCR3 | PLSCR3 Entrez, Source | phospholipid scramblase 3 | 12584 | -0.124 | -0.3659 | Yes |
| 95 | NCDN | NCDN Entrez, Source | neurochondrin | 12607 | -0.126 | -0.3532 | Yes |
| 96 | DOK1 | DOK1 Entrez, Source | docking protein 1, 62kDa (downstream of tyrosine kinase 1) | 12620 | -0.127 | -0.3396 | Yes |
| 97 | CALD1 | CALD1 Entrez, Source | caldesmon 1 | 12735 | -0.139 | -0.3323 | Yes |
| 98 | PXN | PXN Entrez, Source | paxillin | 12950 | -0.176 | -0.3285 | Yes |
| 99 | TPM1 | TPM1 Entrez, Source | tropomyosin 1 (alpha) | 12980 | -0.185 | -0.3096 | Yes |
| 100 | GCNT3 | GCNT3 Entrez, Source | glucosaminyl (N-acetyl) transferase 3, mucin type | 13051 | -0.207 | -0.2912 | Yes |
| 101 | ETS1 | ETS1 Entrez, Source | v-ets erythroblastosis virus E26 oncogene homolog 1 (avian) | 13063 | -0.214 | -0.2676 | Yes |
| 102 | GPM6A | GPM6A Entrez, Source | glycoprotein M6A | 13085 | -0.224 | -0.2436 | Yes |
| 103 | TYRO3 | TYRO3 Entrez, Source | TYRO3 protein tyrosine kinase | 13214 | -0.309 | -0.2181 | Yes |
| 104 | SCHIP1 | SCHIP1 Entrez, Source | schwannomin interacting protein 1 | 13215 | -0.309 | -0.1828 | Yes |
| 105 | LOX | LOX Entrez, Source | lysyl oxidase | 13251 | -0.352 | -0.1453 | Yes |
| 106 | MMP11 | MMP11 Entrez, Source | matrix metallopeptidase 11 (stromelysin 3) | 13286 | -0.422 | -0.0997 | Yes |
| 107 | STMN1 | STMN1 Entrez, Source | stathmin 1/oncoprotein 18 | 13294 | -0.447 | -0.0491 | Yes |
| 108 | PPP2R3A | PPP2R3A Entrez, Source | protein phosphatase 2 (formerly 2A), regulatory subunit B'', alpha | 13298 | -0.461 | 0.0033 | Yes |

