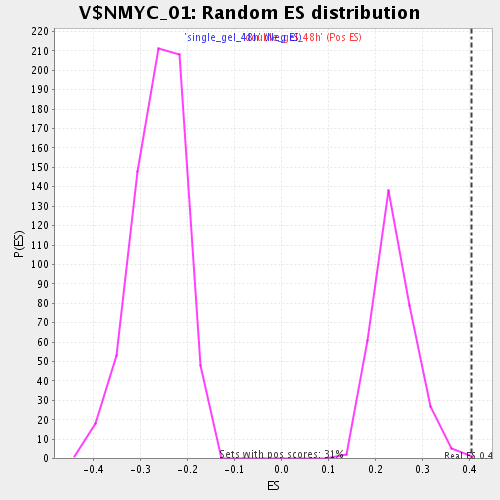
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_48h_versus_single_gel_48h.class.cls #double_gel_48h_versus_single_gel_48h_repos |
| Phenotype | class.cls#double_gel_48h_versus_single_gel_48h_repos |
| Upregulated in class | double_gel_48h |
| GeneSet | V$NMYC_01 |
| Enrichment Score (ES) | 0.40328562 |
| Normalized Enrichment Score (NES) | 1.6817528 |
| Nominal p-value | 0.0031948881 |
| FDR q-value | 0.061929967 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | ELOVL6 | ELOVL6 Entrez, Source | ELOVL family member 6, elongation of long chain fatty acids (FEN1/Elo2, SUR4/Elo3-like, yeast) | 29 | 0.313 | 0.0397 | Yes |
| 2 | USP2 | USP2 Entrez, Source | ubiquitin specific peptidase 2 | 42 | 0.293 | 0.0781 | Yes |
| 3 | ABCA1 | ABCA1 Entrez, Source | ATP-binding cassette, sub-family A (ABC1), member 1 | 71 | 0.251 | 0.1096 | Yes |
| 4 | GADD45G | GADD45G Entrez, Source | growth arrest and DNA-damage-inducible, gamma | 160 | 0.189 | 0.1283 | Yes |
| 5 | CHRM1 | CHRM1 Entrez, Source | cholinergic receptor, muscarinic 1 | 169 | 0.184 | 0.1524 | Yes |
| 6 | SC5DL | SC5DL Entrez, Source | sterol-C5-desaturase (ERG3 delta-5-desaturase homolog, fungal)-like | 187 | 0.176 | 0.1746 | Yes |
| 7 | FKBP5 | FKBP5 Entrez, Source | FK506 binding protein 5 | 219 | 0.169 | 0.1948 | Yes |
| 8 | GPC3 | GPC3 Entrez, Source | glypican 3 | 221 | 0.167 | 0.2171 | Yes |
| 9 | POU3F1 | POU3F1 Entrez, Source | POU domain, class 3, transcription factor 1 | 291 | 0.150 | 0.2320 | Yes |
| 10 | RCL1 | RCL1 Entrez, Source | RNA terminal phosphate cyclase-like 1 | 432 | 0.132 | 0.2391 | Yes |
| 11 | GPD1 | GPD1 Entrez, Source | glycerol-3-phosphate dehydrogenase 1 (soluble) | 555 | 0.121 | 0.2461 | Yes |
| 12 | AP1GBP1 | AP1GBP1 Entrez, Source | AP1 gamma subunit binding protein 1 | 560 | 0.121 | 0.2619 | Yes |
| 13 | VGF | VGF Entrez, Source | VGF nerve growth factor inducible | 587 | 0.119 | 0.2759 | Yes |
| 14 | PPAT | PPAT Entrez, Source | phosphoribosyl pyrophosphate amidotransferase | 709 | 0.111 | 0.2816 | Yes |
| 15 | ATAD3A | ATAD3A Entrez, Source | ATPase family, AAA domain containing 3A | 721 | 0.111 | 0.2956 | Yes |
| 16 | CEBPB | CEBPB Entrez, Source | CCAAT/enhancer binding protein (C/EBP), beta | 732 | 0.110 | 0.3095 | Yes |
| 17 | DLC1 | DLC1 Entrez, Source | deleted in liver cancer 1 | 960 | 0.099 | 0.3056 | Yes |
| 18 | FGF11 | FGF11 Entrez, Source | fibroblast growth factor 11 | 1029 | 0.096 | 0.3134 | Yes |
| 19 | TESK2 | TESK2 Entrez, Source | testis-specific kinase 2 | 1137 | 0.092 | 0.3175 | Yes |
| 20 | PPARGC1B | PPARGC1B Entrez, Source | peroxisome proliferative activated receptor, gamma, coactivator 1, beta | 1166 | 0.091 | 0.3275 | Yes |
| 21 | TIMM10 | TIMM10 Entrez, Source | translocase of inner mitochondrial membrane 10 homolog (yeast) | 1198 | 0.090 | 0.3372 | Yes |
| 22 | DUSP4 | DUSP4 Entrez, Source | dual specificity phosphatase 4 | 1225 | 0.089 | 0.3471 | Yes |
| 23 | HRSP12 | HRSP12 Entrez, Source | heat-responsive protein 12 | 1267 | 0.087 | 0.3557 | Yes |
| 24 | SLC20A1 | SLC20A1 Entrez, Source | solute carrier family 20 (phosphate transporter), member 1 | 1308 | 0.086 | 0.3641 | Yes |
| 25 | MTHFD1 | MTHFD1 Entrez, Source | methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase | 1392 | 0.083 | 0.3690 | Yes |
| 26 | SEPT3 | SEPT3 Entrez, Source | septin 3 | 1429 | 0.082 | 0.3772 | Yes |
| 27 | ADCY3 | ADCY3 Entrez, Source | adenylate cyclase 3 | 1611 | 0.077 | 0.3738 | Yes |
| 28 | WBP2 | WBP2 Entrez, Source | WW domain binding protein 2 | 1725 | 0.074 | 0.3751 | Yes |
| 29 | DNAJB9 | DNAJB9 Entrez, Source | DnaJ (Hsp40) homolog, subfamily B, member 9 | 1749 | 0.073 | 0.3831 | Yes |
| 30 | ATP5G1 | ATP5G1 Entrez, Source | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C1 (subunit 9) | 1822 | 0.072 | 0.3873 | Yes |
| 31 | PA2G4 | PA2G4 Entrez, Source | proliferation-associated 2G4, 38kDa | 1958 | 0.069 | 0.3863 | Yes |
| 32 | LIG3 | LIG3 Entrez, Source | ligase III, DNA, ATP-dependent | 2002 | 0.068 | 0.3922 | Yes |
| 33 | PRKCE | PRKCE Entrez, Source | protein kinase C, epsilon | 2008 | 0.068 | 0.4009 | Yes |
| 34 | TIMM9 | TIMM9 Entrez, Source | translocase of inner mitochondrial membrane 9 homolog (yeast) | 2118 | 0.065 | 0.4014 | Yes |
| 35 | BMP2K | BMP2K Entrez, Source | BMP2 inducible kinase | 2207 | 0.064 | 0.4033 | Yes |
| 36 | NEF3 | NEF3 Entrez, Source | neurofilament 3 (150kDa medium) | 2564 | 0.057 | 0.3840 | No |
| 37 | HMOX1 | HMOX1 Entrez, Source | heme oxygenase (decycling) 1 | 2730 | 0.055 | 0.3789 | No |
| 38 | HMGN2 | HMGN2 Entrez, Source | high-mobility group nucleosomal binding domain 2 | 3017 | 0.050 | 0.3639 | No |
| 39 | GCSH | GCSH Entrez, Source | glycine cleavage system protein H (aminomethyl carrier) | 3126 | 0.048 | 0.3622 | No |
| 40 | DLX1 | DLX1 Entrez, Source | distal-less homeobox 1 | 3150 | 0.048 | 0.3669 | No |
| 41 | NOL5A | NOL5A Entrez, Source | nucleolar protein 5A (56kDa with KKE/D repeat) | 3346 | 0.045 | 0.3582 | No |
| 42 | IPO13 | IPO13 Entrez, Source | importin 13 | 3500 | 0.043 | 0.3523 | No |
| 43 | LAP3 | LAP3 Entrez, Source | leucine aminopeptidase 3 | 3603 | 0.042 | 0.3502 | No |
| 44 | RXRB | RXRB Entrez, Source | retinoid X receptor, beta | 3669 | 0.041 | 0.3507 | No |
| 45 | MYST3 | MYST3 Entrez, Source | MYST histone acetyltransferase (monocytic leukemia) 3 | 3748 | 0.040 | 0.3502 | No |
| 46 | GRIN2A | GRIN2A Entrez, Source | glutamate receptor, ionotropic, N-methyl D-aspartate 2A | 3905 | 0.038 | 0.3434 | No |
| 47 | TOP1 | TOP1 Entrez, Source | topoisomerase (DNA) I | 3919 | 0.038 | 0.3475 | No |
| 48 | FBL | FBL Entrez, Source | fibrillarin | 4004 | 0.036 | 0.3460 | No |
| 49 | SLC12A5 | SLC12A5 Entrez, Source | solute carrier family 12, (potassium-chloride transporter) member 5 | 4112 | 0.035 | 0.3426 | No |
| 50 | CACNA1D | CACNA1D Entrez, Source | calcium channel, voltage-dependent, L type, alpha 1D subunit | 4301 | 0.033 | 0.3327 | No |
| 51 | PLA2G4A | PLA2G4A Entrez, Source | phospholipase A2, group IVA (cytosolic, calcium-dependent) | 4307 | 0.033 | 0.3367 | No |
| 52 | BCL2 | BCL2 Entrez, Source | B-cell CLL/lymphoma 2 | 4328 | 0.032 | 0.3395 | No |
| 53 | PFKFB3 | PFKFB3 Entrez, Source | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 | 4397 | 0.031 | 0.3385 | No |
| 54 | TEF | TEF Entrez, Source | thyrotrophic embryonic factor | 4623 | 0.028 | 0.3253 | No |
| 55 | RUNX2 | RUNX2 Entrez, Source | runt-related transcription factor 2 | 4704 | 0.027 | 0.3230 | No |
| 56 | GJA1 | GJA1 Entrez, Source | gap junction protein, alpha 1, 43kDa (connexin 43) | 4783 | 0.027 | 0.3206 | No |
| 57 | SGTB | SGTB Entrez, Source | small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta | 4819 | 0.026 | 0.3214 | No |
| 58 | MAPKAPK3 | MAPKAPK3 Entrez, Source | mitogen-activated protein kinase-activated protein kinase 3 | 4926 | 0.025 | 0.3167 | No |
| 59 | NPM1 | NPM1 Entrez, Source | nucleophosmin (nucleolar phosphoprotein B23, numatrin) | 4979 | 0.024 | 0.3160 | No |
| 60 | NUP153 | NUP153 Entrez, Source | nucleoporin 153kDa | 5047 | 0.023 | 0.3141 | No |
| 61 | HYAL2 | HYAL2 Entrez, Source | hyaluronoglucosaminidase 2 | 5215 | 0.021 | 0.3043 | No |
| 62 | HSPE1 | HSPE1 Entrez, Source | heat shock 10kDa protein 1 (chaperonin 10) | 5218 | 0.021 | 0.3070 | No |
| 63 | NRXN1 | NRXN1 Entrez, Source | neurexin 1 | 5290 | 0.020 | 0.3043 | No |
| 64 | NDUFS1 | NDUFS1 Entrez, Source | NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) | 5421 | 0.019 | 0.2971 | No |
| 65 | ADPRHL1 | ADPRHL1 Entrez, Source | ADP-ribosylhydrolase like 1 | 5477 | 0.019 | 0.2954 | No |
| 66 | ANGPT2 | ANGPT2 Entrez, Source | angiopoietin 2 | 5512 | 0.018 | 0.2952 | No |
| 67 | HSPD1 | HSPD1 Entrez, Source | heat shock 60kDa protein 1 (chaperonin) | 5662 | 0.016 | 0.2862 | No |
| 68 | EIF4G1 | EIF4G1 Entrez, Source | eukaryotic translation initiation factor 4 gamma, 1 | 5763 | 0.015 | 0.2807 | No |
| 69 | PLAG1 | PLAG1 Entrez, Source | pleiomorphic adenoma gene 1 | 5765 | 0.015 | 0.2826 | No |
| 70 | HTF9C | HTF9C Entrez, Source | - | 6047 | 0.012 | 0.2630 | No |
| 71 | NR1D1 | NR1D1 Entrez, Source | nuclear receptor subfamily 1, group D, member 1 | 6058 | 0.012 | 0.2639 | No |
| 72 | COL2A1 | COL2A1 Entrez, Source | collagen, type II, alpha 1 (primary osteoarthritis, spondyloepiphyseal dysplasia, congenital) | 6063 | 0.012 | 0.2652 | No |
| 73 | FGF6 | FGF6 Entrez, Source | fibroblast growth factor 6 | 6199 | 0.011 | 0.2565 | No |
| 74 | RAMP2 | RAMP2 Entrez, Source | receptor (calcitonin) activity modifying protein 2 | 6398 | 0.009 | 0.2427 | No |
| 75 | PABPC1 | PABPC1 Entrez, Source | poly(A) binding protein, cytoplasmic 1 | 6465 | 0.008 | 0.2387 | No |
| 76 | HPCA | HPCA Entrez, Source | hippocalcin | 6474 | 0.008 | 0.2392 | No |
| 77 | ARF6 | ARF6 Entrez, Source | ADP-ribosylation factor 6 | 6899 | 0.003 | 0.2075 | No |
| 78 | PDK2 | PDK2 Entrez, Source | pyruvate dehydrogenase kinase, isozyme 2 | 7313 | -0.002 | 0.1765 | No |
| 79 | CLCN2 | CLCN2 Entrez, Source | chloride channel 2 | 7332 | -0.002 | 0.1753 | No |
| 80 | NEUROD2 | NEUROD2 Entrez, Source | neurogenic differentiation 2 | 7475 | -0.003 | 0.1651 | No |
| 81 | SLC38A5 | SLC38A5 Entrez, Source | solute carrier family 38, member 5 | 7548 | -0.004 | 0.1602 | No |
| 82 | HRH3 | HRH3 Entrez, Source | histamine receptor H3 | 7593 | -0.005 | 0.1575 | No |
| 83 | RPL13A | RPL13A Entrez, Source | ribosomal protein L13a | 7679 | -0.006 | 0.1518 | No |
| 84 | NXPH1 | NXPH1 Entrez, Source | neurexophilin 1 | 7716 | -0.006 | 0.1499 | No |
| 85 | TLE3 | TLE3 Entrez, Source | transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila) | 7718 | -0.006 | 0.1506 | No |
| 86 | APOA5 | APOA5 Entrez, Source | apolipoprotein A-V | 7763 | -0.007 | 0.1482 | No |
| 87 | RBBP6 | RBBP6 Entrez, Source | retinoblastoma binding protein 6 | 7799 | -0.007 | 0.1465 | No |
| 88 | PABPC4 | PABPC4 Entrez, Source | poly(A) binding protein, cytoplasmic 4 (inducible form) | 8106 | -0.011 | 0.1248 | No |
| 89 | BARHL1 | BARHL1 Entrez, Source | BarH-like 1 (Drosophila) | 8283 | -0.013 | 0.1132 | No |
| 90 | XTP3TPA | XTP3TPA Entrez, Source | - | 8400 | -0.014 | 0.1063 | No |
| 91 | RPL22 | RPL22 Entrez, Source | ribosomal protein L22 | 8557 | -0.016 | 0.0967 | No |
| 92 | DNMT3A | DNMT3A Entrez, Source | DNA (cytosine-5-)-methyltransferase 3 alpha | 8594 | -0.017 | 0.0962 | No |
| 93 | RAB2 | RAB2 Entrez, Source | RAB2, member RAS oncogene family | 8693 | -0.018 | 0.0912 | No |
| 94 | ARHGAP20 | ARHGAP20 Entrez, Source | Rho GTPase activating protein 20 | 8786 | -0.019 | 0.0868 | No |
| 95 | VPS16 | VPS16 Entrez, Source | vacuolar protein sorting 16 (yeast) | 8885 | -0.020 | 0.0821 | No |
| 96 | HBP1 | HBP1 Entrez, Source | HMG-box transcription factor 1 | 8915 | -0.021 | 0.0826 | No |
| 97 | STMN4 | STMN4 Entrez, Source | stathmin-like 4 | 8943 | -0.021 | 0.0834 | No |
| 98 | RBM3 | RBM3 Entrez, Source | RNA binding motif (RNP1, RRM) protein 3 | 9354 | -0.027 | 0.0560 | No |
| 99 | U2AF2 | U2AF2 Entrez, Source | U2 small nuclear RNA auxiliary factor 2 | 9410 | -0.028 | 0.0555 | No |
| 100 | STAT3 | STAT3 Entrez, Source | signal transducer and activator of transcription 3 (acute-phase response factor) | 9416 | -0.028 | 0.0589 | No |
| 101 | ZFP91 | ZFP91 Entrez, Source | zinc finger protein 91 homolog (mouse) | 9444 | -0.028 | 0.0606 | No |
| 102 | SH3KBP1 | SH3KBP1 Entrez, Source | SH3-domain kinase binding protein 1 | 9559 | -0.030 | 0.0560 | No |
| 103 | GPS1 | GPS1 Entrez, Source | G protein pathway suppressor 1 | 9588 | -0.030 | 0.0579 | No |
| 104 | SLC9A5 | SLC9A5 Entrez, Source | solute carrier family 9 (sodium/hydrogen exchanger), member 5 | 9614 | -0.031 | 0.0601 | No |
| 105 | ANXA6 | ANXA6 Entrez, Source | annexin A6 | 9885 | -0.035 | 0.0443 | No |
| 106 | PFN1 | PFN1 Entrez, Source | profilin 1 | 10319 | -0.042 | 0.0172 | No |
| 107 | SCYL1 | SCYL1 Entrez, Source | SCY1-like 1 (S. cerevisiae) | 10331 | -0.042 | 0.0220 | No |
| 108 | MAX | MAX Entrez, Source | MYC associated factor X | 10590 | -0.047 | 0.0087 | No |
| 109 | GADD45B | GADD45B Entrez, Source | growth arrest and DNA-damage-inducible, beta | 10614 | -0.047 | 0.0133 | No |
| 110 | GIT1 | GIT1 Entrez, Source | G protein-coupled receptor kinase interactor 1 | 10969 | -0.055 | -0.0061 | No |
| 111 | CUGBP1 | CUGBP1 Entrez, Source | CUG triplet repeat, RNA binding protein 1 | 11120 | -0.058 | -0.0096 | No |
| 112 | ADAM10 | ADAM10 Entrez, Source | ADAM metallopeptidase domain 10 | 11154 | -0.059 | -0.0042 | No |
| 113 | PRDX4 | PRDX4 Entrez, Source | peroxiredoxin 4 | 11241 | -0.061 | -0.0025 | No |
| 114 | PLAGL1 | PLAGL1 Entrez, Source | pleiomorphic adenoma gene-like 1 | 11247 | -0.061 | 0.0053 | No |
| 115 | EIF4B | EIF4B Entrez, Source | eukaryotic translation initiation factor 4B | 11325 | -0.063 | 0.0079 | No |
| 116 | presenilin 2 (Alzheimer disease 4) | PSEN2 Entrez, Source | 11726 | -0.076 | -0.0122 | No | |
| 117 | CTBP2 | CTBP2 Entrez, Source | C-terminal binding protein 2 | 11872 | -0.082 | -0.0122 | No |
| 118 | GRK6 | GRK6 Entrez, Source | G protein-coupled receptor kinase 6 | 11990 | -0.086 | -0.0095 | No |
| 119 | G6PC3 | G6PC3 Entrez, Source | glucose 6 phosphatase, catalytic, 3 | 12117 | -0.092 | -0.0067 | No |
| 120 | NRAS | NRAS Entrez, Source | neuroblastoma RAS viral (v-ras) oncogene homolog | 12153 | -0.094 | 0.0033 | No |
| 121 | RAB30 | RAB30 Entrez, Source | RAB30, member RAS oncogene family | 12489 | -0.117 | -0.0064 | No |
| 122 | WEE1 | WEE1 Entrez, Source | WEE1 homolog (S. pombe) | 12774 | -0.145 | -0.0085 | No |
| 123 | CTSF | CTSF Entrez, Source | cathepsin F | 12816 | -0.150 | 0.0084 | No |
| 124 | LMNB1 | LMNB1 Entrez, Source | lamin B1 | 13104 | -0.234 | 0.0180 | No |