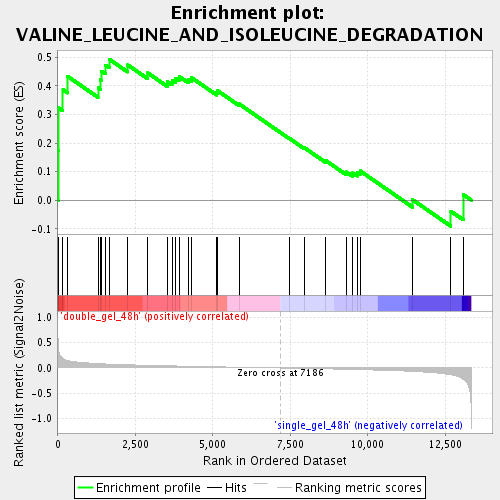
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_48h_versus_single_gel_48h.class.cls #double_gel_48h_versus_single_gel_48h_repos |
| Phenotype | class.cls#double_gel_48h_versus_single_gel_48h_repos |
| Upregulated in class | double_gel_48h |
| GeneSet | VALINE_LEUCINE_AND_ISOLEUCINE_DEGRADATION |
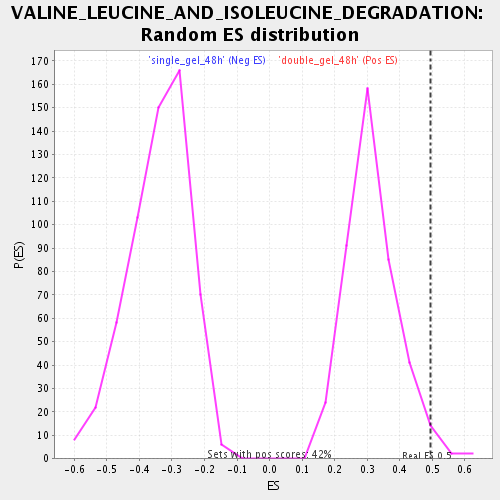
| Enrichment Score (ES) | 0.49363628 |
| Normalized Enrichment Score (NES) | 1.5613897 |
| Nominal p-value | 0.026378896 |
| FDR q-value | 0.11615451 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | EHHADH | EHHADH Entrez, Source | enoyl-Coenzyme A, hydratase/3-hydroxyacyl Coenzyme A dehydrogenase | 6 | 0.447 | 0.1757 | Yes |
| 2 | ACAT2 | ACAT2 Entrez, Source | acetyl-Coenzyme A acetyltransferase 2 (acetoacetyl Coenzyme A thiolase) | 14 | 0.377 | 0.3238 | Yes |
| 3 | ACAA2 | ACAA2 Entrez, Source | acetyl-Coenzyme A acyltransferase 2 (mitochondrial 3-oxoacyl-Coenzyme A thiolase) | 163 | 0.187 | 0.3865 | Yes |
| 4 | ALDH6A1 | ALDH6A1 Entrez, Source | aldehyde dehydrogenase 6 family, member A1 | 314 | 0.148 | 0.4335 | Yes |
| 5 | ACAT1 | ACAT1 Entrez, Source | acetyl-Coenzyme A acetyltransferase 1 (acetoacetyl Coenzyme A thiolase) | 1295 | 0.086 | 0.3938 | Yes |
| 6 | AOX1 | AOX1 Entrez, Source | aldehyde oxidase 1 | 1365 | 0.084 | 0.4216 | Yes |
| 7 | HMGCL | HMGCL Entrez, Source | 3-hydroxymethyl-3-methylglutaryl-Coenzyme A lyase (hydroxymethylglutaricaciduria) | 1394 | 0.083 | 0.4522 | Yes |
| 8 | ECHS1 | ECHS1 Entrez, Source | enoyl Coenzyme A hydratase, short chain, 1, mitochondrial | 1528 | 0.079 | 0.4731 | Yes |
| 9 | ALDH1A1 | ALDH1A1 Entrez, Source | aldehyde dehydrogenase 1 family, member A1 | 1653 | 0.076 | 0.4936 | Yes |
| 10 | BCKDHA | BCKDHA Entrez, Source | branched chain keto acid dehydrogenase E1, alpha polypeptide | 2252 | 0.063 | 0.4735 | No |
| 11 | ALDH1A2 | ALDH1A2 Entrez, Source | aldehyde dehydrogenase 1 family, member A2 | 2896 | 0.052 | 0.4456 | No |
| 12 | ACADSB | ACADSB Entrez, Source | acyl-Coenzyme A dehydrogenase, short/branched chain | 3526 | 0.043 | 0.4152 | No |
| 13 | SDS | SDS Entrez, Source | serine dehydratase | 3684 | 0.041 | 0.4194 | No |
| 14 | HIBADH | HIBADH Entrez, Source | 3-hydroxyisobutyrate dehydrogenase | 3797 | 0.039 | 0.4265 | No |
| 15 | BCKDHB | BCKDHB Entrez, Source | branched chain keto acid dehydrogenase E1, beta polypeptide (maple syrup urine disease) | 3912 | 0.038 | 0.4328 | No |
| 16 | PCCB | PCCB Entrez, Source | propionyl Coenzyme A carboxylase, beta polypeptide | 4212 | 0.034 | 0.4236 | No |
| 17 | HADHB | HADHB Entrez, Source | hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (trifunctional protein), beta subunit | 4313 | 0.032 | 0.4288 | No |
| 18 | MCCC1 | MCCC1 Entrez, Source | methylcrotonoyl-Coenzyme A carboxylase 1 (alpha) | 5123 | 0.022 | 0.3769 | No |
| 19 | HADHA | HADHA Entrez, Source | hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (trifunctional protein), alpha subunit | 5156 | 0.022 | 0.3831 | No |
| 20 | ACADM | ACADM Entrez, Source | acyl-Coenzyme A dehydrogenase, C-4 to C-12 straight chain | 5845 | 0.014 | 0.3371 | No |
| 21 | ALDH3A1 | ALDH3A1 Entrez, Source | aldehyde dehydrogenase 3 family, memberA1 | 7461 | -0.003 | 0.2171 | No |
| 22 | ACADS | ACADS Entrez, Source | acyl-Coenzyme A dehydrogenase, C-2 to C-3 short chain | 7949 | -0.009 | 0.1840 | No |
| 23 | ALDH2 | ALDH2 Entrez, Source | aldehyde dehydrogenase 2 family (mitochondrial) | 8630 | -0.017 | 0.1397 | No |
| 24 | ALDH9A1 | ALDH9A1 Entrez, Source | aldehyde dehydrogenase 9 family, member A1 | 9309 | -0.026 | 0.0991 | No |
| 25 | ALDH3A2 | ALDH3A2 Entrez, Source | aldehyde dehydrogenase 3 family, member A2 | 9511 | -0.029 | 0.0954 | No |
| 26 | ACADL | ACADL Entrez, Source | acyl-Coenzyme A dehydrogenase, long chain | 9656 | -0.031 | 0.0970 | No |
| 27 | MCCC2 | MCCC2 Entrez, Source | methylcrotonoyl-Coenzyme A carboxylase 2 (beta) | 9753 | -0.033 | 0.1027 | No |
| 28 | IVD | IVD Entrez, Source | isovaleryl Coenzyme A dehydrogenase | 11444 | -0.066 | 0.0019 | No |
| 29 | ALDH1B1 | ALDH1B1 Entrez, Source | aldehyde dehydrogenase 1 family, member B1 | 12682 | -0.133 | -0.0386 | No |
| 30 | BCAT1 | BCAT1 Entrez, Source | branched chain aminotransferase 1, cytosolic | 13084 | -0.224 | 0.0194 | No |