Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_72h_versus_single_gel_72h.class.cls #double_gel_72h_versus_single_gel_72h_repos |
| Phenotype | class.cls#double_gel_72h_versus_single_gel_72h_repos |
| Upregulated in class | single_gel_72h |
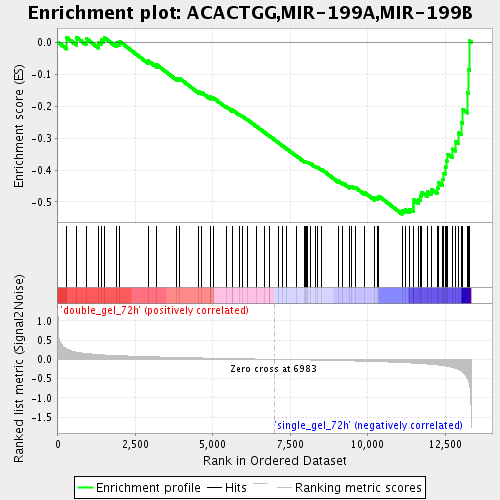
| GeneSet | ACACTGG,MIR-199A,MIR-199B |
| Enrichment Score (ES) | -0.5379153 |
| Normalized Enrichment Score (NES) | -1.7798806 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.040465906 |
| FWER p-Value | 0.992 |

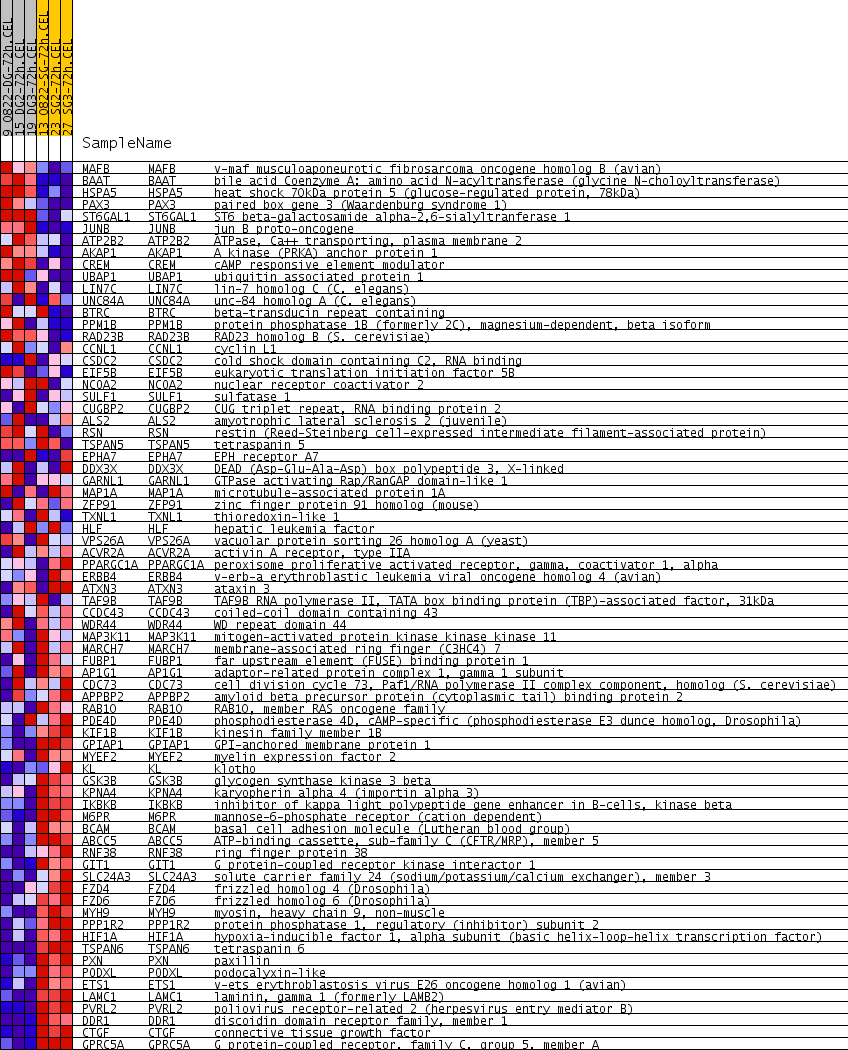
| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | MAFB | MAFB Entrez, Source | v-maf musculoaponeurotic fibrosarcoma oncogene homolog B (avian) | 285 | 0.273 | 0.0143 | No |
| 2 | BAAT | BAAT Entrez, Source | bile acid Coenzyme A: amino acid N-acyltransferase (glycine N-choloyltransferase) | 597 | 0.186 | 0.0153 | No |
| 3 | HSPA5 | HSPA5 Entrez, Source | heat shock 70kDa protein 5 (glucose-regulated protein, 78kDa) | 909 | 0.152 | 0.0118 | No |
| 4 | PAX3 | PAX3 Entrez, Source | paired box gene 3 (Waardenburg syndrome 1) | 1312 | 0.123 | -0.0024 | No |
| 5 | ST6GAL1 | ST6GAL1 Entrez, Source | ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 | 1393 | 0.119 | 0.0072 | No |
| 6 | JUNB | JUNB Entrez, Source | jun B proto-oncogene | 1487 | 0.114 | 0.0151 | No |
| 7 | ATP2B2 | ATP2B2 Entrez, Source | ATPase, Ca++ transporting, plasma membrane 2 | 1881 | 0.098 | -0.0017 | No |
| 8 | AKAP1 | AKAP1 Entrez, Source | A kinase (PRKA) anchor protein 1 | 2000 | 0.094 | 0.0018 | No |
| 9 | CREM | CREM Entrez, Source | cAMP responsive element modulator | 2911 | 0.069 | -0.0578 | No |
| 10 | UBAP1 | UBAP1 Entrez, Source | ubiquitin associated protein 1 | 3192 | 0.062 | -0.0708 | No |
| 11 | LIN7C | LIN7C Entrez, Source | lin-7 homolog C (C. elegans) | 3835 | 0.049 | -0.1127 | No |
| 12 | UNC84A | UNC84A Entrez, Source | unc-84 homolog A (C. elegans) | 3935 | 0.048 | -0.1139 | No |
| 13 | BTRC | BTRC Entrez, Source | beta-transducin repeat containing | 4551 | 0.037 | -0.1554 | No |
| 14 | PPM1B | PPM1B Entrez, Source | protein phosphatase 1B (formerly 2C), magnesium-dependent, beta isoform | 4644 | 0.035 | -0.1577 | No |
| 15 | RAD23B | RAD23B Entrez, Source | RAD23 homolog B (S. cerevisiae) | 4914 | 0.031 | -0.1740 | No |
| 16 | CCNL1 | CCNL1 Entrez, Source | cyclin L1 | 4932 | 0.030 | -0.1713 | No |
| 17 | CSDC2 | CSDC2 Entrez, Source | cold shock domain containing C2, RNA binding | 5025 | 0.029 | -0.1745 | No |
| 18 | EIF5B | EIF5B Entrez, Source | eukaryotic translation initiation factor 5B | 5435 | 0.023 | -0.2023 | No |
| 19 | NCOA2 | NCOA2 Entrez, Source | nuclear receptor coactivator 2 | 5627 | 0.020 | -0.2141 | No |
| 20 | SULF1 | SULF1 Entrez, Source | sulfatase 1 | 5633 | 0.020 | -0.2119 | No |
| 21 | CUGBP2 | CUGBP2 Entrez, Source | CUG triplet repeat, RNA binding protein 2 | 5853 | 0.017 | -0.2262 | No |
| 22 | ALS2 | ALS2 Entrez, Source | amyotrophic lateral sclerosis 2 (juvenile) | 5959 | 0.015 | -0.2322 | No |
| 23 | RSN | RSN Entrez, Source | restin (Reed-Steinberg cell-expressed intermediate filament-associated protein) | 6102 | 0.013 | -0.2413 | No |
| 24 | TSPAN5 | TSPAN5 Entrez, Source | tetraspanin 5 | 6410 | 0.008 | -0.2634 | No |
| 25 | EPHA7 | EPHA7 Entrez, Source | EPH receptor A7 | 6653 | 0.005 | -0.2810 | No |
| 26 | DDX3X | DDX3X Entrez, Source | DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, X-linked | 6818 | 0.002 | -0.2931 | No |
| 27 | GARNL1 | GARNL1 Entrez, Source | GTPase activating Rap/RanGAP domain-like 1 | 6825 | 0.002 | -0.2932 | No |
| 28 | MAP1A | MAP1A Entrez, Source | microtubule-associated protein 1A | 7116 | -0.002 | -0.3148 | No |
| 29 | ZFP91 | ZFP91 Entrez, Source | zinc finger protein 91 homolog (mouse) | 7235 | -0.004 | -0.3233 | No |
| 30 | TXNL1 | TXNL1 Entrez, Source | thioredoxin-like 1 | 7392 | -0.006 | -0.3342 | No |
| 31 | HLF | HLF Entrez, Source | hepatic leukemia factor | 7695 | -0.010 | -0.3557 | No |
| 32 | VPS26A | VPS26A Entrez, Source | vacuolar protein sorting 26 homolog A (yeast) | 7970 | -0.015 | -0.3744 | No |
| 33 | ACVR2A | ACVR2A Entrez, Source | activin A receptor, type IIA | 7984 | -0.015 | -0.3735 | No |
| 34 | PPARGC1A | PPARGC1A Entrez, Source | peroxisome proliferative activated receptor, gamma, coactivator 1, alpha | 8036 | -0.015 | -0.3753 | No |
| 35 | ERBB4 | ERBB4 Entrez, Source | v-erb-a erythroblastic leukemia viral oncogene homolog 4 (avian) | 8064 | -0.016 | -0.3753 | No |
| 36 | ATXN3 | ATXN3 Entrez, Source | ataxin 3 | 8145 | -0.017 | -0.3791 | No |
| 37 | TAF9B | TAF9B Entrez, Source | TAF9B RNA polymerase II, TATA box binding protein (TBP)-associated factor, 31kDa | 8303 | -0.020 | -0.3883 | No |
| 38 | CCDC43 | CCDC43 Entrez, Source | coiled-coil domain containing 43 | 8372 | -0.021 | -0.3907 | No |
| 39 | WDR44 | WDR44 Entrez, Source | WD repeat domain 44 | 8519 | -0.023 | -0.3986 | No |
| 40 | MAP3K11 | MAP3K11 Entrez, Source | mitogen-activated protein kinase kinase kinase 11 | 9043 | -0.033 | -0.4337 | No |
| 41 | MARCH7 | MARCH7 Entrez, Source | membrane-associated ring finger (C3HC4) 7 | 9196 | -0.036 | -0.4405 | No |
| 42 | FUBP1 | FUBP1 Entrez, Source | far upstream element (FUSE) binding protein 1 | 9417 | -0.040 | -0.4519 | No |
| 43 | AP1G1 | AP1G1 Entrez, Source | adaptor-related protein complex 1, gamma 1 subunit | 9466 | -0.041 | -0.4501 | No |
| 44 | CDC73 | CDC73 Entrez, Source | cell division cycle 73, Paf1/RNA polymerase II complex component, homolog (S. cerevisiae) | 9588 | -0.043 | -0.4536 | No |
| 45 | APPBP2 | APPBP2 Entrez, Source | amyloid beta precursor protein (cytoplasmic tail) binding protein 2 | 9900 | -0.050 | -0.4704 | No |
| 46 | RAB10 | RAB10 Entrez, Source | RAB10, member RAS oncogene family | 10219 | -0.057 | -0.4869 | No |
| 47 | PDE4D | PDE4D Entrez, Source | phosphodiesterase 4D, cAMP-specific (phosphodiesterase E3 dunce homolog, Drosophila) | 10306 | -0.059 | -0.4856 | No |
| 48 | KIF1B | KIF1B Entrez, Source | kinesin family member 1B | 10358 | -0.060 | -0.4815 | No |
| 49 | GPIAP1 | GPIAP1 Entrez, Source | GPI-anchored membrane protein 1 | 11107 | -0.082 | -0.5272 | Yes |
| 50 | MYEF2 | MYEF2 Entrez, Source | myelin expression factor 2 | 11204 | -0.086 | -0.5231 | Yes |
| 51 | KL | KL Entrez, Source | klotho | 11354 | -0.093 | -0.5222 | Yes |
| 52 | GSK3B | GSK3B Entrez, Source | glycogen synthase kinase 3 beta | 11474 | -0.098 | -0.5183 | Yes |
| 53 | KPNA4 | KPNA4 Entrez, Source | karyopherin alpha 4 (importin alpha 3) | 11478 | -0.099 | -0.5056 | Yes |
| 54 | IKBKB | IKBKB Entrez, Source | inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta | 11490 | -0.099 | -0.4934 | Yes |
| 55 | M6PR | M6PR Entrez, Source | mannose-6-phosphate receptor (cation dependent) | 11645 | -0.106 | -0.4911 | Yes |
| 56 | BCAM | BCAM Entrez, Source | basal cell adhesion molecule (Lutheran blood group) | 11705 | -0.109 | -0.4812 | Yes |
| 57 | ABCC5 | ABCC5 Entrez, Source | ATP-binding cassette, sub-family C (CFTR/MRP), member 5 | 11735 | -0.111 | -0.4689 | Yes |
| 58 | RNF38 | RNF38 Entrez, Source | ring finger protein 38 | 11931 | -0.120 | -0.4679 | Yes |
| 59 | GIT1 | GIT1 Entrez, Source | G protein-coupled receptor kinase interactor 1 | 12064 | -0.129 | -0.4609 | Yes |
| 60 | SLC24A3 | SLC24A3 Entrez, Source | solute carrier family 24 (sodium/potassium/calcium exchanger), member 3 | 12251 | -0.144 | -0.4561 | Yes |
| 61 | FZD4 | FZD4 Entrez, Source | frizzled homolog 4 (Drosophila) | 12275 | -0.146 | -0.4387 | Yes |
| 62 | FZD6 | FZD6 Entrez, Source | frizzled homolog 6 (Drosophila) | 12417 | -0.160 | -0.4284 | Yes |
| 63 | MYH9 | MYH9 Entrez, Source | myosin, heavy chain 9, non-muscle | 12456 | -0.165 | -0.4097 | Yes |
| 64 | PPP1R2 | PPP1R2 Entrez, Source | protein phosphatase 1, regulatory (inhibitor) subunit 2 | 12496 | -0.170 | -0.3904 | Yes |
| 65 | HIF1A | HIF1A Entrez, Source | hypoxia-inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) | 12528 | -0.175 | -0.3698 | Yes |
| 66 | TSPAN6 | TSPAN6 Entrez, Source | tetraspanin 6 | 12584 | -0.182 | -0.3502 | Yes |
| 67 | PXN | PXN Entrez, Source | paxillin | 12740 | -0.211 | -0.3342 | Yes |
| 68 | PODXL | PODXL Entrez, Source | podocalyxin-like | 12834 | -0.233 | -0.3107 | Yes |
| 69 | ETS1 | ETS1 Entrez, Source | v-ets erythroblastosis virus E26 oncogene homolog 1 (avian) | 12930 | -0.262 | -0.2835 | Yes |
| 70 | LAMC1 | LAMC1 Entrez, Source | laminin, gamma 1 (formerly LAMB2) | 13018 | -0.300 | -0.2508 | Yes |
| 71 | PVRL2 | PVRL2 Entrez, Source | poliovirus receptor-related 2 (herpesvirus entry mediator B) | 13071 | -0.333 | -0.2111 | Yes |
| 72 | DDR1 | DDR1 Entrez, Source | discoidin domain receptor family, member 1 | 13214 | -0.489 | -0.1577 | Yes |
| 73 | CTGF | CTGF Entrez, Source | connective tissue growth factor | 13260 | -0.571 | -0.0862 | Yes |
| 74 | GPRC5A | GPRC5A Entrez, Source | G protein-coupled receptor, family C, group 5, member A | 13289 | -0.705 | 0.0040 | Yes |