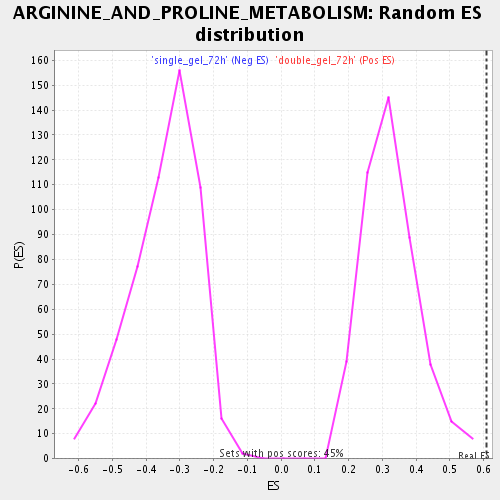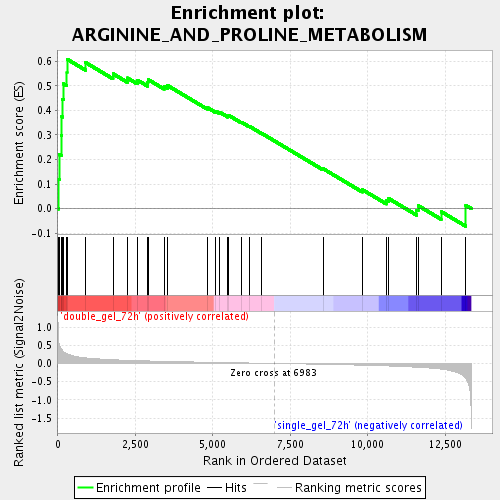
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_72h_versus_single_gel_72h.class.cls #double_gel_72h_versus_single_gel_72h_repos |
| Phenotype | class.cls#double_gel_72h_versus_single_gel_72h_repos |
| Upregulated in class | double_gel_72h |
| GeneSet | ARGININE_AND_PROLINE_METABOLISM |
| Enrichment Score (ES) | 0.6083736 |
| Normalized Enrichment Score (NES) | 1.8687881 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.008967573 |
| FWER p-Value | 0.62 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | ABP1 | ABP1 Entrez, Source | amiloride binding protein 1 (amine oxidase (copper-containing)) | 24 | 0.594 | 0.1186 | Yes |
| 2 | OTC | OTC Entrez, Source | ornithine carbamoyltransferase | 44 | 0.506 | 0.2198 | Yes |
| 3 | GOT1 | GOT1 Entrez, Source | glutamic-oxaloacetic transaminase 1, soluble (aspartate aminotransferase 1) | 110 | 0.403 | 0.2966 | Yes |
| 4 | AGMAT | AGMAT Entrez, Source | agmatine ureohydrolase (agmatinase) | 124 | 0.386 | 0.3740 | Yes |
| 5 | ARG1 | ARG1 Entrez, Source | arginase, liver | 145 | 0.358 | 0.4452 | Yes |
| 6 | ASL | ASL Entrez, Source | argininosuccinate lyase | 183 | 0.323 | 0.5081 | Yes |
| 7 | CPS1 | CPS1 Entrez, Source | carbamoyl-phosphate synthetase 1, mitochondrial | 292 | 0.268 | 0.5543 | Yes |
| 8 | GAMT | GAMT Entrez, Source | guanidinoacetate N-methyltransferase | 295 | 0.267 | 0.6084 | Yes |
| 9 | OAT | OAT Entrez, Source | ornithine aminotransferase (gyrate atrophy) | 893 | 0.154 | 0.5947 | No |
| 10 | MAOB | MAOB Entrez, Source | monoamine oxidase B | 1779 | 0.102 | 0.5489 | No |
| 11 | ALDH1A1 | ALDH1A1 Entrez, Source | aldehyde dehydrogenase 1 family, member A1 | 2239 | 0.087 | 0.5320 | No |
| 12 | AMD1 | AMD1 Entrez, Source | adenosylmethionine decarboxylase 1 | 2566 | 0.078 | 0.5232 | No |
| 13 | AOC3 | AOC3 Entrez, Source | amine oxidase, copper containing 3 (vascular adhesion protein 1) | 2897 | 0.069 | 0.5125 | No |
| 14 | NOS1 | NOS1 Entrez, Source | nitric oxide synthase 1 (neuronal) | 2908 | 0.069 | 0.5256 | No |
| 15 | MAOA | MAOA Entrez, Source | monoamine oxidase A | 3427 | 0.057 | 0.4984 | No |
| 16 | P4HB | P4HB Entrez, Source | procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), beta polypeptide | 3536 | 0.055 | 0.5015 | No |
| 17 | ALDH3A2 | ALDH3A2 Entrez, Source | aldehyde dehydrogenase 3 family, member A2 | 4819 | 0.032 | 0.4117 | No |
| 18 | GLUD1 | GLUD1 Entrez, Source | glutamate dehydrogenase 1 | 5087 | 0.028 | 0.3972 | No |
| 19 | ALDH1B1 | ALDH1B1 Entrez, Source | aldehyde dehydrogenase 1 family, member B1 | 5218 | 0.026 | 0.3928 | No |
| 20 | ODC1 | ODC1 Entrez, Source | ornithine decarboxylase 1 | 5487 | 0.022 | 0.3771 | No |
| 21 | ALDH1A2 | ALDH1A2 Entrez, Source | aldehyde dehydrogenase 1 family, member A2 | 5520 | 0.021 | 0.3790 | No |
| 22 | GOT2 | GOT2 Entrez, Source | glutamic-oxaloacetic transaminase 2, mitochondrial (aspartate aminotransferase 2) | 5916 | 0.015 | 0.3525 | No |
| 23 | ALDH2 | ALDH2 Entrez, Source | aldehyde dehydrogenase 2 family (mitochondrial) | 6175 | 0.012 | 0.3355 | No |
| 24 | ARG2 | ARG2 Entrez, Source | arginase, type II | 6580 | 0.006 | 0.3062 | No |
| 25 | CKM | CKM Entrez, Source | creatine kinase, muscle | 8557 | -0.024 | 0.1627 | No |
| 26 | ALDH3A1 | ALDH3A1 Entrez, Source | aldehyde dehydrogenase 3 family, memberA1 | 9824 | -0.048 | 0.0774 | No |
| 27 | CKB | CKB Entrez, Source | creatine kinase, brain | 10604 | -0.067 | 0.0324 | No |
| 28 | ALDH9A1 | ALDH9A1 Entrez, Source | aldehyde dehydrogenase 9 family, member A1 | 10674 | -0.069 | 0.0413 | No |
| 29 | GATM | GATM Entrez, Source | glycine amidinotransferase (L-arginine:glycine amidinotransferase) | 11580 | -0.103 | -0.0058 | No |
| 30 | P4HA1 | P4HA1 Entrez, Source | procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), alpha polypeptide I | 11627 | -0.106 | 0.0122 | No |
| 31 | NOS3 | NOS3 Entrez, Source | nitric oxide synthase 3 (endothelial cell) | 12369 | -0.155 | -0.0121 | No |
| 32 | SMS | SMS Entrez, Source | spermine synthase | 13163 | -0.420 | 0.0134 | No |