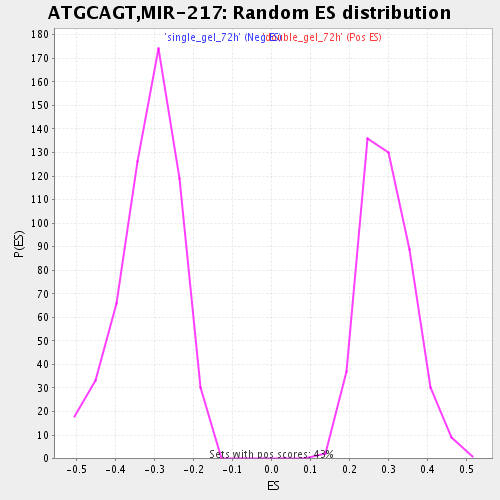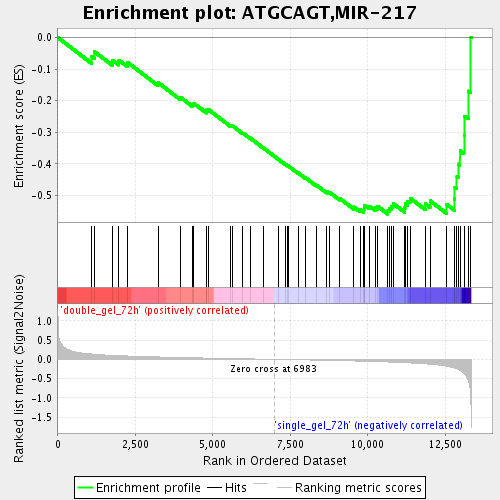
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_72h_versus_single_gel_72h.class.cls #double_gel_72h_versus_single_gel_72h_repos |
| Phenotype | class.cls#double_gel_72h_versus_single_gel_72h_repos |
| Upregulated in class | single_gel_72h |
| GeneSet | ATGCAGT,MIR-217 |
| Enrichment Score (ES) | -0.5586669 |
| Normalized Enrichment Score (NES) | -1.7826817 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.03982514 |
| FWER p-Value | 0.989 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | JOSD3 | JOSD3 Entrez, Source | Josephin domain containing 3 | 1089 | 0.138 | -0.0595 | No |
| 2 | SLC31A1 | SLC31A1 Entrez, Source | solute carrier family 31 (copper transporters), member 1 | 1173 | 0.132 | -0.0442 | No |
| 3 | FBN2 | FBN2 Entrez, Source | fibrillin 2 (congenital contractural arachnodactyly) | 1754 | 0.103 | -0.0710 | No |
| 4 | CHN2 | CHN2 Entrez, Source | chimerin (chimaerin) 2 | 1966 | 0.095 | -0.0714 | No |
| 5 | NR4A2 | NR4A2 Entrez, Source | nuclear receptor subfamily 4, group A, member 2 | 2245 | 0.086 | -0.0783 | No |
| 6 | POLG | POLG Entrez, Source | polymerase (DNA directed), gamma | 3231 | 0.062 | -0.1423 | No |
| 7 | STX1A | STX1A Entrez, Source | syntaxin 1A (brain) | 3958 | 0.047 | -0.1893 | No |
| 8 | GRIK2 | GRIK2 Entrez, Source | glutamate receptor, ionotropic, kainate 2 | 4329 | 0.041 | -0.2105 | No |
| 9 | TBC1D15 | TBC1D15 Entrez, Source | TBC1 domain family, member 15 | 4388 | 0.040 | -0.2084 | No |
| 10 | SEPT4 | SEPT4 Entrez, Source | septin 4 | 4795 | 0.033 | -0.2336 | No |
| 11 | STAT5B | STAT5B Entrez, Source | signal transducer and activator of transcription 5B | 4806 | 0.032 | -0.2291 | No |
| 12 | NFIA | NFIA Entrez, Source | nuclear factor I/A | 4861 | 0.032 | -0.2280 | No |
| 13 | ST18 | ST18 Entrez, Source | suppression of tumorigenicity 18 (breast carcinoma) (zinc finger protein) | 5573 | 0.021 | -0.2781 | No |
| 14 | MAMDC2 | MAMDC2 Entrez, Source | MAM domain containing 2 | 5639 | 0.020 | -0.2798 | No |
| 15 | PCNA | PCNA Entrez, Source | proliferating cell nuclear antigen | 5968 | 0.015 | -0.3021 | No |
| 16 | BCL11A | BCL11A Entrez, Source | B-cell CLL/lymphoma 11A (zinc finger protein) | 6200 | 0.011 | -0.3176 | No |
| 17 | STRBP | STRBP Entrez, Source | spermatid perinuclear RNA binding protein | 6648 | 0.005 | -0.3505 | No |
| 18 | NOVA1 | NOVA1 Entrez, Source | neuro-oncological ventral antigen 1 | 7115 | -0.002 | -0.3853 | No |
| 19 | KCNH5 | KCNH5 Entrez, Source | potassium voltage-gated channel, subfamily H (eag-related), member 5 | 7342 | -0.005 | -0.4014 | No |
| 20 | ATP1B1 | ATP1B1 Entrez, Source | ATPase, Na+/K+ transporting, beta 1 polypeptide | 7404 | -0.006 | -0.4051 | No |
| 21 | GATAD2B | GATAD2B Entrez, Source | GATA zinc finger domain containing 2B | 7430 | -0.006 | -0.4059 | No |
| 22 | CUL5 | CUL5 Entrez, Source | cullin 5 | 7748 | -0.011 | -0.4279 | No |
| 23 | ACVR2A | ACVR2A Entrez, Source | activin A receptor, type IIA | 7984 | -0.015 | -0.4432 | No |
| 24 | DYRK1A | DYRK1A Entrez, Source | dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A | 8340 | -0.020 | -0.4666 | No |
| 25 | OGT | OGT Entrez, Source | O-linked N-acetylglucosamine (GlcNAc) transferase (UDP-N-acetylglucosamine:polypeptide-N-acetylglucosaminyl transferase) | 8678 | -0.026 | -0.4877 | No |
| 26 | EIF4A2 | EIF4A2 Entrez, Source | eukaryotic translation initiation factor 4A, isoform 2 | 8760 | -0.027 | -0.4893 | No |
| 27 | GDI2 | GDI2 Entrez, Source | GDP dissociation inhibitor 2 | 9090 | -0.034 | -0.5085 | No |
| 28 | SLC38A2 | SLC38A2 Entrez, Source | solute carrier family 38, member 2 | 9549 | -0.043 | -0.5360 | No |
| 29 | MAPK1 | MAPK1 Entrez, Source | mitogen-activated protein kinase 1 | 9755 | -0.047 | -0.5438 | No |
| 30 | UBXD8 | UBXD8 Entrez, Source | UBX domain containing 8 | 9877 | -0.049 | -0.5449 | No |
| 31 | KRAS | KRAS Entrez, Source | v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog | 9882 | -0.049 | -0.5371 | No |
| 32 | APPBP2 | APPBP2 Entrez, Source | amyloid beta precursor protein (cytoplasmic tail) binding protein 2 | 9900 | -0.050 | -0.5302 | No |
| 33 | FN1 | FN1 Entrez, Source | fibronectin 1 | 10062 | -0.053 | -0.5337 | No |
| 34 | CAPN3 | CAPN3 Entrez, Source | calpain 3, (p94) | 10237 | -0.058 | -0.5374 | No |
| 35 | UBL3 | UBL3 Entrez, Source | ubiquitin-like 3 | 10310 | -0.059 | -0.5331 | No |
| 36 | MTPN | MTPN Entrez, Source | myotrophin | 10651 | -0.068 | -0.5475 | Yes |
| 37 | DLGAP2 | DLGAP2 Entrez, Source | discs, large (Drosophila) homolog-associated protein 2 | 10712 | -0.070 | -0.5406 | Yes |
| 38 | SOX11 | SOX11 Entrez, Source | SRY (sex determining region Y)-box 11 | 10773 | -0.072 | -0.5334 | Yes |
| 39 | KCNA3 | KCNA3 Entrez, Source | potassium voltage-gated channel, shaker-related subfamily, member 3 | 10815 | -0.073 | -0.5246 | Yes |
| 40 | NDEL1 | NDEL1 Entrez, Source | nudE nuclear distribution gene E homolog like 1 (A. nidulans) | 11189 | -0.085 | -0.5387 | Yes |
| 41 | MYEF2 | MYEF2 Entrez, Source | myelin expression factor 2 | 11204 | -0.086 | -0.5258 | Yes |
| 42 | NEUROD1 | NEUROD1 Entrez, Source | neurogenic differentiation 1 | 11288 | -0.090 | -0.5174 | Yes |
| 43 | YWHAG | YWHAG Entrez, Source | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide | 11367 | -0.093 | -0.5080 | Yes |
| 44 | FBXO11 | FBXO11 Entrez, Source | F-box protein 11 | 11862 | -0.117 | -0.5261 | Yes |
| 45 | TACC2 | TACC2 Entrez, Source | transforming, acidic coiled-coil containing protein 2 | 12013 | -0.126 | -0.5168 | Yes |
| 46 | DYNC1LI2 | DYNC1LI2 Entrez, Source | dynein, cytoplasmic 1, light intermediate chain 2 | 12539 | -0.177 | -0.5275 | Yes |
| 47 | NIPBL | NIPBL Entrez, Source | Nipped-B homolog (Drosophila) | 12794 | -0.223 | -0.5103 | Yes |
| 48 | MAF | MAF Entrez, Source | v-maf musculoaponeurotic fibrosarcoma oncogene homolog (avian) | 12812 | -0.228 | -0.4743 | Yes |
| 49 | STX6 | STX6 Entrez, Source | syntaxin 6 | 12878 | -0.244 | -0.4393 | Yes |
| 50 | TMSB4X | TMSB4X Entrez, Source | thymosin, beta 4, X-linked | 12920 | -0.259 | -0.4001 | Yes |
| 51 | ADAMTS5 | ADAMTS5 Entrez, Source | ADAM metallopeptidase with thrombospondin type 1 motif, 5 (aggrecanase-2) | 12977 | -0.282 | -0.3583 | Yes |
| 52 | AKAP2 | AKAP2 Entrez, Source | A kinase (PRKA) anchor protein 2 | 13107 | -0.362 | -0.3089 | Yes |
| 53 | TACC1 | TACC1 Entrez, Source | transforming, acidic coiled-coil containing protein 1 | 13123 | -0.377 | -0.2485 | Yes |
| 54 | GPM6A | GPM6A Entrez, Source | glycoprotein M6A | 13245 | -0.543 | -0.1691 | Yes |
| 55 | MSN | MSN Entrez, Source | moesin | 13329 | -1.080 | 0.0010 | Yes |