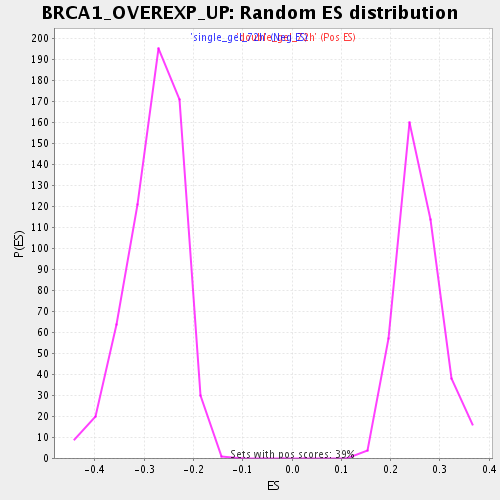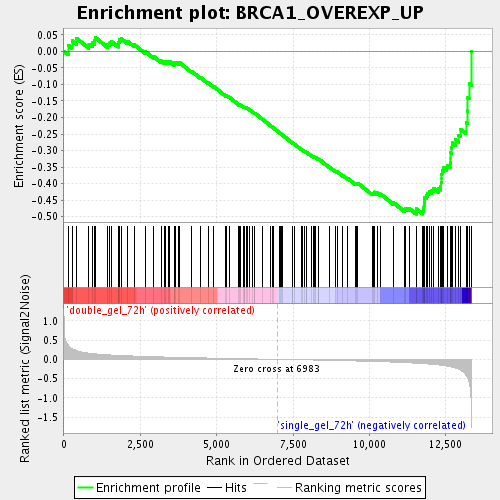
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_72h_versus_single_gel_72h.class.cls #double_gel_72h_versus_single_gel_72h_repos |
| Phenotype | class.cls#double_gel_72h_versus_single_gel_72h_repos |
| Upregulated in class | single_gel_72h |
| GeneSet | BRCA1_OVEREXP_UP |
| Enrichment Score (ES) | -0.4922369 |
| Normalized Enrichment Score (NES) | -1.7649136 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.042279128 |
| FWER p-Value | 0.999 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | ENPP2 | ENPP2 Entrez, Source | ectonucleotide pyrophosphatase/phosphodiesterase 2 (autotaxin) | 143 | 0.363 | 0.0186 | No |
| 2 | ID4 | ID4 Entrez, Source | inhibitor of DNA binding 4, dominant negative helix-loop-helix protein | 265 | 0.281 | 0.0322 | No |
| 3 | SEPHS2 | SEPHS2 Entrez, Source | selenophosphate synthetase 2 | 404 | 0.230 | 0.0403 | No |
| 4 | HAGH | HAGH Entrez, Source | hydroxyacylglutathione hydrolase | 819 | 0.161 | 0.0220 | No |
| 5 | LTB4DH | LTB4DH Entrez, Source | leukotriene B4 12-hydroxydehydrogenase | 931 | 0.151 | 0.0258 | No |
| 6 | LAMA3 | LAMA3 Entrez, Source | laminin, alpha 3 | 1001 | 0.144 | 0.0323 | No |
| 7 | ATP6V0A1 | ATP6V0A1 Entrez, Source | ATPase, H+ transporting, lysosomal V0 subunit a1 | 1017 | 0.143 | 0.0428 | No |
| 8 | SSR1 | SSR1 Entrez, Source | signal sequence receptor, alpha (translocon-associated protein alpha) | 1437 | 0.116 | 0.0205 | No |
| 9 | IDH1 | IDH1 Entrez, Source | isocitrate dehydrogenase 1 (NADP+), soluble | 1494 | 0.114 | 0.0255 | No |
| 10 | TUBB3 | TUBB3 Entrez, Source | tubulin, beta 3 | 1557 | 0.110 | 0.0297 | No |
| 11 | PPP1R1A | PPP1R1A Entrez, Source | protein phosphatase 1, regulatory (inhibitor) subunit 1A | 1778 | 0.102 | 0.0213 | No |
| 12 | RABGGTB | RABGGTB Entrez, Source | Rab geranylgeranyltransferase, beta subunit | 1794 | 0.101 | 0.0284 | No |
| 13 | NDUFS1 | NDUFS1 Entrez, Source | NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) | 1817 | 0.101 | 0.0349 | No |
| 14 | PCBD1 | PCBD1 Entrez, Source | pterin-4 alpha-carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha (TCF1) | 1870 | 0.099 | 0.0389 | No |
| 15 | KDELR2 | KDELR2 Entrez, Source | KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2 | 2086 | 0.091 | 0.0301 | No |
| 16 | WWP1 | WWP1 Entrez, Source | WW domain containing E3 ubiquitin protein ligase 1 | 2306 | 0.085 | 0.0204 | No |
| 17 | HIST2H2AA3 | HIST2H2AA3 Entrez, Source | histone cluster 2, H2aa3 | 2660 | 0.075 | -0.0003 | No |
| 18 | CSDA | CSDA Entrez, Source | cold shock domain protein A | 2927 | 0.068 | -0.0149 | No |
| 19 | P2RX4 | P2RX4 Entrez, Source | purinergic receptor P2X, ligand-gated ion channel, 4 | 3178 | 0.063 | -0.0287 | No |
| 20 | TIMM17A | TIMM17A Entrez, Source | translocase of inner mitochondrial membrane 17 homolog A (yeast) | 3292 | 0.060 | -0.0324 | No |
| 21 | ABCD3 | ABCD3 Entrez, Source | ATP-binding cassette, sub-family D (ALD), member 3 | 3314 | 0.060 | -0.0291 | No |
| 22 | STAT1 | STAT1 Entrez, Source | signal transducer and activator of transcription 1, 91kDa | 3416 | 0.058 | -0.0321 | No |
| 23 | KCNK1 | KCNK1 Entrez, Source | potassium channel, subfamily K, member 1 | 3465 | 0.057 | -0.0312 | No |
| 24 | ALDH7A1 | ALDH7A1 Entrez, Source | aldehyde dehydrogenase 7 family, member A1 | 3614 | 0.053 | -0.0380 | No |
| 25 | TAF11 | TAF11 Entrez, Source | TAF11 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 28kDa | 3629 | 0.053 | -0.0348 | No |
| 26 | TSNAX | TSNAX Entrez, Source | translin-associated factor X | 3667 | 0.053 | -0.0333 | No |
| 27 | SMC1A | SMC1A Entrez, Source | structural maintenance of chromosomes 1A | 3744 | 0.051 | -0.0349 | No |
| 28 | COPS8 | COPS8 Entrez, Source | COP9 constitutive photomorphogenic homolog subunit 8 (Arabidopsis) | 3778 | 0.050 | -0.0334 | No |
| 29 | SEPP1 | SEPP1 Entrez, Source | selenoprotein P, plasma, 1 | 4188 | 0.043 | -0.0608 | No |
| 30 | FSTL1 | FSTL1 Entrez, Source | follistatin-like 1 | 4464 | 0.038 | -0.0785 | No |
| 31 | PPP6C | PPP6C Entrez, Source | protein phosphatase 6, catalytic subunit | 4715 | 0.034 | -0.0947 | No |
| 32 | CD9 | CD9 Entrez, Source | CD9 molecule | 4896 | 0.031 | -0.1058 | No |
| 33 | NFYC | NFYC Entrez, Source | nuclear transcription factor Y, gamma | 5302 | 0.025 | -0.1344 | No |
| 34 | ARF6 | ARF6 Entrez, Source | ADP-ribosylation factor 6 | 5321 | 0.024 | -0.1338 | No |
| 35 | ADD3 | ADD3 Entrez, Source | adducin 3 (gamma) | 5403 | 0.023 | -0.1380 | No |
| 36 | TFPI2 | TFPI2 Entrez, Source | tissue factor pathway inhibitor 2 | 5729 | 0.018 | -0.1611 | No |
| 37 | SUCLG1 | SUCLG1 Entrez, Source | succinate-CoA ligase, GDP-forming, alpha subunit | 5738 | 0.018 | -0.1603 | No |
| 38 | SDHC | SDHC Entrez, Source | succinate dehydrogenase complex, subunit C, integral membrane protein, 15kDa | 5789 | 0.018 | -0.1626 | No |
| 39 | TEGT | TEGT Entrez, Source | testis enhanced gene transcript (BAX inhibitor 1) | 5892 | 0.016 | -0.1691 | No |
| 40 | SFRS5 | SFRS5 Entrez, Source | splicing factor, arginine/serine-rich 5 | 5904 | 0.016 | -0.1686 | No |
| 41 | ADCYAP1 | ADCYAP1 Entrez, Source | adenylate cyclase activating polypeptide 1 (pituitary) | 5921 | 0.015 | -0.1686 | No |
| 42 | TM9SF2 | TM9SF2 Entrez, Source | transmembrane 9 superfamily member 2 | 5966 | 0.015 | -0.1707 | No |
| 43 | GUCY1B3 | GUCY1B3 Entrez, Source | guanylate cyclase 1, soluble, beta 3 | 5991 | 0.014 | -0.1714 | No |
| 44 | AGPS | AGPS Entrez, Source | alkylglycerone phosphate synthase | 6079 | 0.013 | -0.1769 | No |
| 45 | TMED10 | TMED10 Entrez, Source | transmembrane emp24-like trafficking protein 10 (yeast) | 6185 | 0.012 | -0.1839 | No |
| 46 | TM9SF1 | TM9SF1 Entrez, Source | transmembrane 9 superfamily member 1 | 6248 | 0.010 | -0.1878 | No |
| 47 | GALNT3 | GALNT3 Entrez, Source | UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 3 (GalNAc-T3) | 6251 | 0.010 | -0.1871 | No |
| 48 | RAB6A | RAB6A Entrez, Source | RAB6A, member RAS oncogene family | 6512 | 0.006 | -0.2062 | No |
| 49 | ID2 | ID2 Entrez, Source | inhibitor of DNA binding 2, dominant negative helix-loop-helix protein | 6773 | 0.003 | -0.2257 | No |
| 50 | DDX3X | DDX3X Entrez, Source | DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, X-linked | 6818 | 0.002 | -0.2288 | No |
| 51 | TSG101 | TSG101 Entrez, Source | tumor susceptibility gene 101 | 6868 | 0.002 | -0.2323 | No |
| 52 | MAP2K4 | MAP2K4 Entrez, Source | mitogen-activated protein kinase kinase 4 | 7050 | -0.001 | -0.2460 | No |
| 53 | COPS2 | COPS2 Entrez, Source | COP9 constitutive photomorphogenic homolog subunit 2 (Arabidopsis) | 7084 | -0.002 | -0.2483 | No |
| 54 | RAB1A | RAB1A Entrez, Source | RAB1A, member RAS oncogene family | 7133 | -0.002 | -0.2518 | No |
| 55 | TMED2 | TMED2 Entrez, Source | transmembrane emp24 domain trafficking protein 2 | 7149 | -0.002 | -0.2527 | No |
| 56 | ZNF148 | ZNF148 Entrez, Source | zinc finger protein 148 | 7480 | -0.007 | -0.2771 | No |
| 57 | CREB1 | CREB1 Entrez, Source | cAMP responsive element binding protein 1 | 7495 | -0.007 | -0.2776 | No |
| 58 | UBE2D3 | UBE2D3 Entrez, Source | ubiquitin-conjugating enzyme E2D 3 (UBC4/5 homolog, yeast) | 7556 | -0.008 | -0.2814 | No |
| 59 | NID1 | NID1 Entrez, Source | nidogen 1 | 7761 | -0.011 | -0.2959 | No |
| 60 | ACTR1B | ACTR1B Entrez, Source | ARP1 actin-related protein 1 homolog B, centractin beta (yeast) | 7815 | -0.012 | -0.2989 | No |
| 61 | BNIP3 | BNIP3 Entrez, Source | BCL2/adenovirus E1B 19kDa interacting protein 3 | 7875 | -0.013 | -0.3023 | No |
| 62 | EXTL2 | EXTL2 Entrez, Source | exostoses (multiple)-like 2 | 7923 | -0.014 | -0.3048 | No |
| 63 | RAB2 | RAB2 Entrez, Source | RAB2, member RAS oncogene family | 7949 | -0.014 | -0.3055 | No |
| 64 | FRAP1 | FRAP1 Entrez, Source | FK506 binding protein 12-rapamycin associated protein 1 | 8101 | -0.016 | -0.3156 | No |
| 65 | SCAMP1 | SCAMP1 Entrez, Source | secretory carrier membrane protein 1 | 8178 | -0.018 | -0.3199 | No |
| 66 | ARPC1A | ARPC1A Entrez, Source | actin related protein 2/3 complex, subunit 1A, 41kDa | 8191 | -0.018 | -0.3194 | No |
| 67 | GATA3 | GATA3 Entrez, Source | GATA binding protein 3 | 8231 | -0.019 | -0.3208 | No |
| 68 | CDKN1A | CDKN1A Entrez, Source | cyclin-dependent kinase inhibitor 1A (p21, Cip1) | 8319 | -0.020 | -0.3258 | No |
| 69 | PGRMC1 | PGRMC1 Entrez, Source | progesterone receptor membrane component 1 | 8338 | -0.020 | -0.3255 | No |
| 70 | HTATIP | HTATIP Entrez, Source | HIV-1 Tat interacting protein, 60kDa | 8687 | -0.026 | -0.3497 | No |
| 71 | TRAM1 | TRAM1 Entrez, Source | translocation associated membrane protein 1 | 8883 | -0.030 | -0.3620 | No |
| 72 | HMGN3 | HMGN3 Entrez, Source | high mobility group nucleosomal binding domain 3 | 8947 | -0.031 | -0.3643 | No |
| 73 | TOP2A | TOP2A Entrez, Source | topoisomerase (DNA) II alpha 170kDa | 9129 | -0.035 | -0.3752 | No |
| 74 | VAMP3 | VAMP3 Entrez, Source | vesicle-associated membrane protein 3 (cellubrevin) | 9280 | -0.037 | -0.3835 | No |
| 75 | CSNK1A1 | CSNK1A1 Entrez, Source | casein kinase 1, alpha 1 | 9528 | -0.042 | -0.3987 | No |
| 76 | RDX | RDX Entrez, Source | radixin | 9584 | -0.043 | -0.3994 | No |
| 77 | EXOC5 | EXOC5 Entrez, Source | exocyst complex component 5 | 9622 | -0.044 | -0.3987 | No |
| 78 | TITF1 | TITF1 Entrez, Source | thyroid transcription factor 1 | 10088 | -0.054 | -0.4295 | No |
| 79 | FGF9 | FGF9 Entrez, Source | fibroblast growth factor 9 (glia-activating factor) | 10145 | -0.055 | -0.4292 | No |
| 80 | FCGRT | FCGRT Entrez, Source | Fc fragment of IgG, receptor, transporter, alpha | 10155 | -0.056 | -0.4254 | No |
| 81 | PRKAR1A | PRKAR1A Entrez, Source | protein kinase, cAMP-dependent, regulatory, type I, alpha (tissue specific extinguisher 1) | 10251 | -0.058 | -0.4279 | No |
| 82 | PLD3 | PLD3 Entrez, Source | phospholipase D family, member 3 | 10363 | -0.061 | -0.4314 | No |
| 83 | JAK1 | JAK1 Entrez, Source | Janus kinase 1 (a protein tyrosine kinase) | 10774 | -0.072 | -0.4566 | No |
| 84 | ZFP36L1 | ZFP36L1 Entrez, Source | zinc finger protein 36, C3H type-like 1 | 11158 | -0.084 | -0.4787 | No |
| 85 | DR1 | DR1 Entrez, Source | down-regulator of transcription 1, TBP-binding (negative cofactor 2) | 11186 | -0.085 | -0.4739 | No |
| 86 | MAP2K1 | MAP2K1 Entrez, Source | mitogen-activated protein kinase kinase 1 | 11293 | -0.090 | -0.4746 | No |
| 87 | IGF2R | IGF2R Entrez, Source | insulin-like growth factor 2 receptor | 11527 | -0.101 | -0.4841 | Yes |
| 88 | KIF5B | KIF5B Entrez, Source | kinesin family member 5B | 11531 | -0.101 | -0.4762 | Yes |
| 89 | NEU1 | NEU1 Entrez, Source | sialidase 1 (lysosomal sialidase) | 11743 | -0.111 | -0.4831 | Yes |
| 90 | LUM | LUM Entrez, Source | lumican | 11773 | -0.112 | -0.4762 | Yes |
| 91 | KIF2A | KIF2A Entrez, Source | kinesin heavy chain member 2A | 11783 | -0.113 | -0.4678 | Yes |
| 92 | CXADR | CXADR Entrez, Source | coxsackie virus and adenovirus receptor | 11788 | -0.113 | -0.4590 | Yes |
| 93 | STC1 | STC1 Entrez, Source | stanniocalcin 1 | 11795 | -0.113 | -0.4502 | Yes |
| 94 | CIRBP | CIRBP Entrez, Source | cold inducible RNA binding protein | 11805 | -0.114 | -0.4417 | Yes |
| 95 | SERINC3 | SERINC3 Entrez, Source | serine incorporator 3 | 11857 | -0.117 | -0.4361 | Yes |
| 96 | MFGE8 | MFGE8 Entrez, Source | milk fat globule-EGF factor 8 protein | 11896 | -0.118 | -0.4294 | Yes |
| 97 | DEK | DEK Entrez, Source | DEK oncogene (DNA binding) | 11951 | -0.122 | -0.4236 | Yes |
| 98 | ANXA4 | ANXA4 Entrez, Source | annexin A4 | 12040 | -0.127 | -0.4200 | Yes |
| 99 | NPC2 | NPC2 Entrez, Source | Niemann-Pick disease, type C2 | 12096 | -0.131 | -0.4135 | Yes |
| 100 | PTPN1 | PTPN1 Entrez, Source | protein tyrosine phosphatase, non-receptor type 1 | 12267 | -0.145 | -0.4146 | Yes |
| 101 | ADD1 | ADD1 Entrez, Source | adducin 1 (alpha) | 12334 | -0.152 | -0.4073 | Yes |
| 102 | CALM2 | CALM2 Entrez, Source | calmodulin 2 (phosphorylase kinase, delta) | 12340 | -0.152 | -0.3953 | Yes |
| 103 | WRB | WRB Entrez, Source | tryptophan rich basic protein | 12356 | -0.153 | -0.3841 | Yes |
| 104 | PPP4C | PPP4C Entrez, Source | protein phosphatase 4 (formerly X), catalytic subunit | 12360 | -0.154 | -0.3718 | Yes |
| 105 | ITGB3BP | ITGB3BP Entrez, Source | integrin beta 3 binding protein (beta3-endonexin) | 12374 | -0.155 | -0.3602 | Yes |
| 106 | MAT2A | MAT2A Entrez, Source | methionine adenosyltransferase II, alpha | 12429 | -0.161 | -0.3513 | Yes |
| 107 | ACTA2 | ACTA2 Entrez, Source | actin, alpha 2, smooth muscle, aorta | 12537 | -0.177 | -0.3451 | Yes |
| 108 | PICALM | PICALM Entrez, Source | phosphatidylinositol binding clathrin assembly protein | 12640 | -0.193 | -0.3372 | Yes |
| 109 | PPIC | PPIC Entrez, Source | peptidylprolyl isomerase C (cyclophilin C) | 12659 | -0.196 | -0.3227 | Yes |
| 110 | ANXA2 | ANXA2 Entrez, Source | annexin A2 | 12661 | -0.197 | -0.3068 | Yes |
| 111 | ALCAM | ALCAM Entrez, Source | activated leukocyte cell adhesion molecule | 12680 | -0.201 | -0.2919 | Yes |
| 112 | CD47 | CD47 Entrez, Source | CD47 molecule | 12700 | -0.205 | -0.2768 | Yes |
| 113 | KIF2C | KIF2C Entrez, Source | kinesin family member 2C | 12815 | -0.228 | -0.2669 | Yes |
| 114 | CYBA | CYBA Entrez, Source | cytochrome b-245, alpha polypeptide | 12921 | -0.260 | -0.2538 | Yes |
| 115 | HMMR | HMMR Entrez, Source | hyaluronan-mediated motility receptor (RHAMM) | 12987 | -0.285 | -0.2357 | Yes |
| 116 | PAM | PAM Entrez, Source | peptidylglycine alpha-amidating monooxygenase | 13160 | -0.412 | -0.2153 | Yes |
| 117 | CITED2 | CITED2 Entrez, Source | Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 | 13211 | -0.481 | -0.1802 | Yes |
| 118 | TCEAL1 | TCEAL1 Entrez, Source | transcription elongation factor A (SII)-like 1 | 13218 | -0.493 | -0.1407 | Yes |
| 119 | GNAI1 | GNAI1 Entrez, Source | guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 | 13262 | -0.580 | -0.0970 | Yes |
| 120 | ANXA1 | ANXA1 Entrez, Source | annexin A1 | 13336 | -1.272 | 0.0005 | Yes |