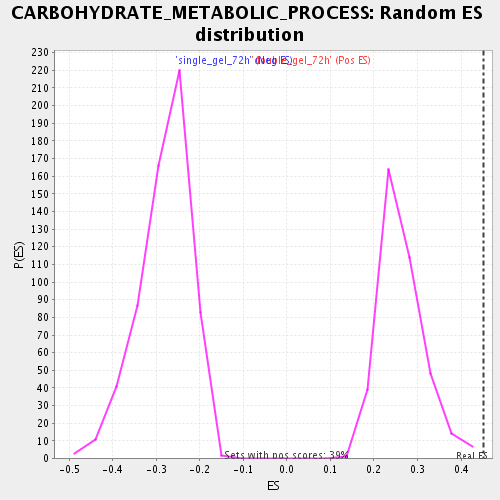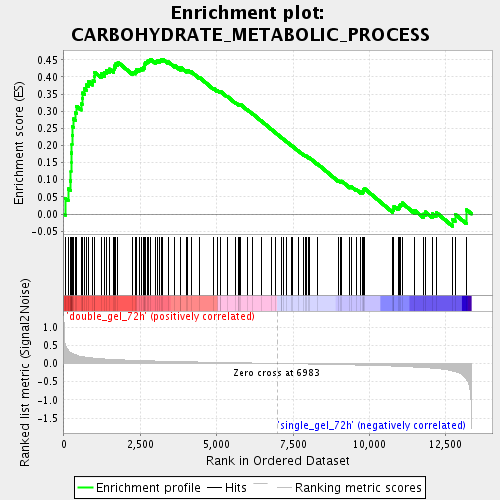
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_72h_versus_single_gel_72h.class.cls #double_gel_72h_versus_single_gel_72h_repos |
| Phenotype | class.cls#double_gel_72h_versus_single_gel_72h_repos |
| Upregulated in class | double_gel_72h |
| GeneSet | CARBOHYDRATE_METABOLIC_PROCESS |
| Enrichment Score (ES) | 0.45187253 |
| Normalized Enrichment Score (NES) | 1.7117298 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.033948127 |
| FWER p-Value | 0.997 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | FBP1 | FBP1 Entrez, Source | fructose-1,6-bisphosphatase 1 | 43 | 0.515 | 0.0458 | Yes |
| 2 | GCK | GCK Entrez, Source | glucokinase (hexokinase 4, maturity onset diabetes of the young 2) | 140 | 0.368 | 0.0735 | Yes |
| 3 | ACLY | ACLY Entrez, Source | ATP citrate lyase | 212 | 0.305 | 0.0971 | Yes |
| 4 | PFKFB1 | PFKFB1 Entrez, Source | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 | 230 | 0.293 | 0.1237 | Yes |
| 5 | MIOX | MIOX Entrez, Source | myo-inositol oxygenase | 234 | 0.291 | 0.1512 | Yes |
| 6 | SLC2A5 | SLC2A5 Entrez, Source | solute carrier family 2 (facilitated glucose/fructose transporter), member 5 | 238 | 0.290 | 0.1785 | Yes |
| 7 | AKR7A2 | AKR7A2 Entrez, Source | aldo-keto reductase family 7, member A2 (aflatoxin aldehyde reductase) | 260 | 0.283 | 0.2039 | Yes |
| 8 | CARKL | CARKL Entrez, Source | carbohydrate kinase-like | 267 | 0.280 | 0.2301 | Yes |
| 9 | ACN9 | ACN9 Entrez, Source | ACN9 homolog (S. cerevisiae) | 284 | 0.273 | 0.2548 | Yes |
| 10 | GYS2 | GYS2 Entrez, Source | glycogen synthase 2 (liver) | 314 | 0.263 | 0.2776 | Yes |
| 11 | SLC25A10 | SLC25A10 Entrez, Source | solute carrier family 25 (mitochondrial carrier; dicarboxylate transporter), member 10 | 378 | 0.240 | 0.2957 | Yes |
| 12 | PDK4 | PDK4 Entrez, Source | pyruvate dehydrogenase kinase, isozyme 4 | 412 | 0.227 | 0.3148 | Yes |
| 13 | PDHB | PDHB Entrez, Source | pyruvate dehydrogenase (lipoamide) beta | 563 | 0.190 | 0.3216 | Yes |
| 14 | GNE | GNE Entrez, Source | glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase | 599 | 0.186 | 0.3366 | Yes |
| 15 | ST8SIA4 | ST8SIA4 Entrez, Source | ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4 | 611 | 0.184 | 0.3534 | Yes |
| 16 | PDK1 | PDK1 Entrez, Source | pyruvate dehydrogenase kinase, isozyme 1 | 673 | 0.176 | 0.3655 | Yes |
| 17 | INSR | INSR Entrez, Source | insulin receptor | 735 | 0.169 | 0.3769 | Yes |
| 18 | ME1 | ME1 Entrez, Source | malic enzyme 1, NADP(+)-dependent, cytosolic | 800 | 0.163 | 0.3876 | Yes |
| 19 | UGDH | UGDH Entrez, Source | UDP-glucose dehydrogenase | 951 | 0.149 | 0.3904 | Yes |
| 20 | XYLB | XYLB Entrez, Source | xylulokinase homolog (H. influenzae) | 995 | 0.145 | 0.4010 | Yes |
| 21 | ALDOB | ALDOB Entrez, Source | aldolase B, fructose-bisphosphate | 1007 | 0.144 | 0.4138 | Yes |
| 22 | ST8SIA1 | ST8SIA1 Entrez, Source | ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1 | 1223 | 0.128 | 0.4098 | Yes |
| 23 | GALNT5 | GALNT5 Entrez, Source | UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5) | 1333 | 0.122 | 0.4132 | Yes |
| 24 | ST6GAL1 | ST6GAL1 Entrez, Source | ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 | 1393 | 0.119 | 0.4201 | Yes |
| 25 | IDH1 | IDH1 Entrez, Source | isocitrate dehydrogenase 1 (NADP+), soluble | 1494 | 0.114 | 0.4233 | Yes |
| 26 | FUT1 | FUT1 Entrez, Source | fucosyltransferase 1 (galactoside 2-alpha-L-fucosyltransferase, H blood group) | 1637 | 0.107 | 0.4228 | Yes |
| 27 | NANP | NANP Entrez, Source | N-acetylneuraminic acid phosphatase | 1648 | 0.107 | 0.4322 | Yes |
| 28 | FUT8 | FUT8 Entrez, Source | fucosyltransferase 8 (alpha (1,6) fucosyltransferase) | 1701 | 0.105 | 0.4382 | Yes |
| 29 | B4GALNT1 | B4GALNT1 Entrez, Source | beta-1,4-N-acetyl-galactosaminyl transferase 1 | 1769 | 0.102 | 0.4429 | Yes |
| 30 | ACO1 | ACO1 Entrez, Source | aconitase 1, soluble | 2259 | 0.086 | 0.4141 | Yes |
| 31 | B3GAT1 | B3GAT1 Entrez, Source | beta-1,3-glucuronyltransferase 1 (glucuronosyltransferase P) | 2334 | 0.084 | 0.4165 | Yes |
| 32 | SLC35A3 | SLC35A3 Entrez, Source | solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3 | 2379 | 0.083 | 0.4211 | Yes |
| 33 | SLC2A2 | SLC2A2 Entrez, Source | solute carrier family 2 (facilitated glucose transporter), member 2 | 2459 | 0.081 | 0.4228 | Yes |
| 34 | SDHD | SDHD Entrez, Source | succinate dehydrogenase complex, subunit D, integral membrane protein | 2529 | 0.079 | 0.4251 | Yes |
| 35 | EPM2A | EPM2A Entrez, Source | epilepsy, progressive myoclonus type 2A, Lafora disease (laforin) | 2594 | 0.077 | 0.4276 | Yes |
| 36 | NDST1 | NDST1 Entrez, Source | N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 | 2645 | 0.075 | 0.4309 | Yes |
| 37 | IDH3B | IDH3B Entrez, Source | isocitrate dehydrogenase 3 (NAD+) beta | 2648 | 0.075 | 0.4379 | Yes |
| 38 | ST6GALNAC6 | ST6GALNAC6 Entrez, Source | ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 | 2672 | 0.074 | 0.4432 | Yes |
| 39 | CHST12 | CHST12 Entrez, Source | carbohydrate (chondroitin 4) sulfotransferase 12 | 2721 | 0.073 | 0.4465 | Yes |
| 40 | SDHA | SDHA Entrez, Source | succinate dehydrogenase complex, subunit A, flavoprotein (Fp) | 2780 | 0.072 | 0.4490 | Yes |
| 41 | PDK2 | PDK2 Entrez, Source | pyruvate dehydrogenase kinase, isozyme 2 | 2834 | 0.071 | 0.4517 | Yes |
| 42 | ST6GAL2 | ST6GAL2 Entrez, Source | ST6 beta-galactosamide alpha-2,6-sialyltranferase 2 | 3005 | 0.067 | 0.4452 | Yes |
| 43 | ATF4 | ATF4 Entrez, Source | activating transcription factor 4 (tax-responsive enhancer element B67) | 3058 | 0.065 | 0.4475 | Yes |
| 44 | ACO2 | ACO2 Entrez, Source | aconitase 2, mitochondrial | 3136 | 0.064 | 0.4477 | Yes |
| 45 | MPDU1 | MPDU1 Entrez, Source | mannose-P-dolichol utilization defect 1 | 3177 | 0.063 | 0.4506 | Yes |
| 46 | CHST3 | CHST3 Entrez, Source | carbohydrate (chondroitin 6) sulfotransferase 3 | 3239 | 0.061 | 0.4519 | Yes |
| 47 | ST3GAL5 | ST3GAL5 Entrez, Source | ST3 beta-galactoside alpha-2,3-sialyltransferase 5 | 3413 | 0.058 | 0.4443 | No |
| 48 | ST8SIA5 | ST8SIA5 Entrez, Source | ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 5 | 3625 | 0.053 | 0.4334 | No |
| 49 | FUT4 | FUT4 Entrez, Source | fucosyltransferase 4 (alpha (1,3) fucosyltransferase, myeloid-specific) | 3809 | 0.050 | 0.4243 | No |
| 50 | FPGT | FPGT Entrez, Source | fucose-1-phosphate guanylyltransferase | 3810 | 0.050 | 0.4290 | No |
| 51 | TALDO1 | TALDO1 Entrez, Source | transaldolase 1 | 4010 | 0.046 | 0.4184 | No |
| 52 | LECT1 | LECT1 Entrez, Source | leukocyte cell derived chemotaxin 1 | 4046 | 0.046 | 0.4201 | No |
| 53 | GAD1 | GAD1 Entrez, Source | glutamate decarboxylase 1 (brain, 67kDa) | 4162 | 0.044 | 0.4155 | No |
| 54 | FUT9 | FUT9 Entrez, Source | fucosyltransferase 9 (alpha (1,3) fucosyltransferase) | 4443 | 0.039 | 0.3980 | No |
| 55 | FBP2 | FBP2 Entrez, Source | fructose-1,6-bisphosphatase 2 | 4892 | 0.031 | 0.3671 | No |
| 56 | ARPP-19 | ARPP-19 Entrez, Source | - | 5020 | 0.029 | 0.3603 | No |
| 57 | POMT1 | POMT1 Entrez, Source | protein-O-mannosyltransferase 1 | 5110 | 0.028 | 0.3562 | No |
| 58 | ADRB3 | ADRB3 Entrez, Source | adrenergic, beta-3-, receptor | 5121 | 0.027 | 0.3580 | No |
| 59 | GALK1 | GALK1 Entrez, Source | galactokinase 1 | 5339 | 0.024 | 0.3439 | No |
| 60 | UGP2 | UGP2 Entrez, Source | UDP-glucose pyrophosphorylase 2 | 5626 | 0.020 | 0.3242 | No |
| 61 | ALDOC | ALDOC Entrez, Source | aldolase C, fructose-bisphosphate | 5718 | 0.018 | 0.3191 | No |
| 62 | PTEN | PTEN Entrez, Source | phosphatase and tensin homolog (mutated in multiple advanced cancers 1) | 5720 | 0.018 | 0.3208 | No |
| 63 | ST3GAL6 | ST3GAL6 Entrez, Source | ST3 beta-galactoside alpha-2,3-sialyltransferase 6 | 5745 | 0.018 | 0.3207 | No |
| 64 | SDHC | SDHC Entrez, Source | succinate dehydrogenase complex, subunit C, integral membrane protein, 15kDa | 5789 | 0.018 | 0.3191 | No |
| 65 | IPPK | IPPK Entrez, Source | inositol 1,3,4,5,6-pentakisphosphate 2-kinase | 6008 | 0.014 | 0.3040 | No |
| 66 | ALDH2 | ALDH2 Entrez, Source | aldehyde dehydrogenase 2 family (mitochondrial) | 6175 | 0.012 | 0.2925 | No |
| 67 | ST8SIA3 | ST8SIA3 Entrez, Source | ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 | 6457 | 0.007 | 0.2720 | No |
| 68 | TFF1 | TFF1 Entrez, Source | trefoil factor 1 (breast cancer, estrogen-inducible sequence expressed in) | 6791 | 0.003 | 0.2471 | No |
| 69 | GAPDHS | GAPDHS Entrez, Source | glyceraldehyde-3-phosphate dehydrogenase, spermatogenic | 6929 | 0.001 | 0.2368 | No |
| 70 | GALT | GALT Entrez, Source | galactose-1-phosphate uridylyltransferase | 7124 | -0.002 | 0.2223 | No |
| 71 | AKR1A1 | AKR1A1 Entrez, Source | aldo-keto reductase family 1, member A1 (aldehyde reductase) | 7127 | -0.002 | 0.2224 | No |
| 72 | B4GALT7 | B4GALT7 Entrez, Source | xylosylprotein beta 1,4-galactosyltransferase, polypeptide 7 (galactosyltransferase I) | 7186 | -0.003 | 0.2183 | No |
| 73 | GBA2 | GBA2 Entrez, Source | glucosidase, beta (bile acid) 2 | 7269 | -0.004 | 0.2124 | No |
| 74 | NEU3 | NEU3 Entrez, Source | sialidase 3 (membrane sialidase) | 7444 | -0.007 | 0.1999 | No |
| 75 | ST8SIA2 | ST8SIA2 Entrez, Source | ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2 | 7470 | -0.007 | 0.1987 | No |
| 76 | B3GAT2 | B3GAT2 Entrez, Source | beta-1,3-glucuronyltransferase 2 (glucuronosyltransferase S) | 7679 | -0.010 | 0.1840 | No |
| 77 | GFPT2 | GFPT2 Entrez, Source | glutamine-fructose-6-phosphate transaminase 2 | 7852 | -0.013 | 0.1722 | No |
| 78 | SLC5A2 | SLC5A2 Entrez, Source | solute carrier family 5 (sodium/glucose cotransporter), member 2 | 7902 | -0.014 | 0.1698 | No |
| 79 | EXTL2 | EXTL2 Entrez, Source | exostoses (multiple)-like 2 | 7923 | -0.014 | 0.1696 | No |
| 80 | PYGM | PYGM Entrez, Source | phosphorylase, glycogen; muscle (McArdle syndrome, glycogen storage disease type V) | 8011 | -0.015 | 0.1645 | No |
| 81 | PPARGC1A | PPARGC1A Entrez, Source | peroxisome proliferative activated receptor, gamma, coactivator 1, alpha | 8036 | -0.015 | 0.1641 | No |
| 82 | LALBA | LALBA Entrez, Source | lactalbumin, alpha- | 8293 | -0.020 | 0.1467 | No |
| 83 | XYLT1 | XYLT1 Entrez, Source | xylosyltransferase I | 8993 | -0.032 | 0.0969 | No |
| 84 | FUT2 | FUT2 Entrez, Source | fucosyltransferase 2 (secretor status included) | 9061 | -0.033 | 0.0950 | No |
| 85 | PIGQ | PIGQ Entrez, Source | phosphatidylinositol glycan anchor biosynthesis, class Q | 9085 | -0.034 | 0.0965 | No |
| 86 | TREH | TREH Entrez, Source | trehalase (brush-border membrane glycoprotein) | 9359 | -0.039 | 0.0795 | No |
| 87 | SLC2A8 | SLC2A8 Entrez, Source | solute carrier family 2, (facilitated glucose transporter) member 8 | 9396 | -0.040 | 0.0806 | No |
| 88 | NAGK | NAGK Entrez, Source | N-acetylglucosamine kinase | 9565 | -0.043 | 0.0720 | No |
| 89 | GALNT7 | GALNT7 Entrez, Source | UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7 (GalNAc-T7) | 9717 | -0.046 | 0.0649 | No |
| 90 | ALDOA | ALDOA Entrez, Source | aldolase A, fructose-bisphosphate | 9779 | -0.047 | 0.0648 | No |
| 91 | SLC2A4 | SLC2A4 Entrez, Source | solute carrier family 2 (facilitated glucose transporter), member 4 | 9794 | -0.048 | 0.0683 | No |
| 92 | GNS | GNS Entrez, Source | glucosamine (N-acetyl)-6-sulfatase (Sanfilippo disease IIID) | 9802 | -0.048 | 0.0724 | No |
| 93 | GMDS | GMDS Entrez, Source | GDP-mannose 4,6-dehydratase | 9847 | -0.049 | 0.0737 | No |
| 94 | PFKL | PFKL Entrez, Source | phosphofructokinase, liver | 10756 | -0.071 | 0.0119 | No |
| 95 | GFPT1 | GFPT1 Entrez, Source | glutamine-fructose-6-phosphate transaminase 1 | 10795 | -0.072 | 0.0159 | No |
| 96 | GAD2 | GAD2 Entrez, Source | glutamate decarboxylase 2 (pancreatic islets and brain, 65kDa) | 10797 | -0.072 | 0.0227 | No |
| 97 | CHST1 | CHST1 Entrez, Source | carbohydrate (keratan sulfate Gal-6) sulfotransferase 1 | 10942 | -0.077 | 0.0191 | No |
| 98 | CHIA | CHIA Entrez, Source | chitinase, acidic | 10976 | -0.078 | 0.0240 | No |
| 99 | GAA | GAA Entrez, Source | glucosidase, alpha; acid (Pompe disease, glycogen storage disease type II) | 11007 | -0.079 | 0.0293 | No |
| 100 | GUSB | GUSB Entrez, Source | glucuronidase, beta | 11065 | -0.081 | 0.0326 | No |
| 101 | GSK3B | GSK3B Entrez, Source | glycogen synthase kinase 3 beta | 11474 | -0.098 | 0.0112 | No |
| 102 | HPSE | HPSE Entrez, Source | heparanase | 11761 | -0.112 | 0.0002 | No |
| 103 | IL17F | IL17F Entrez, Source | interleukin 17F | 11826 | -0.115 | 0.0063 | No |
| 104 | HK1 | HK1 Entrez, Source | hexokinase 1 | 12060 | -0.129 | 0.0009 | No |
| 105 | MGAT1 | MGAT1 Entrez, Source | mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase | 12180 | -0.137 | 0.0050 | No |
| 106 | PYGB | PYGB Entrez, Source | phosphorylase, glycogen; brain | 12718 | -0.208 | -0.0158 | No |
| 107 | PFKM | PFKM Entrez, Source | phosphofructokinase, muscle | 12804 | -0.226 | -0.0007 | No |
| 108 | ST3GAL2 | ST3GAL2 Entrez, Source | ST3 beta-galactoside alpha-2,3-sialyltransferase 2 | 13177 | -0.434 | 0.0125 | No |