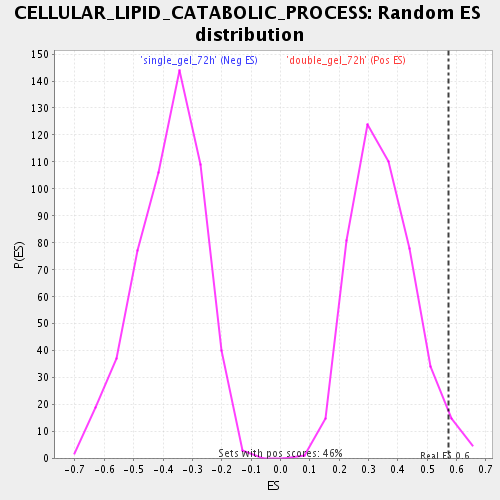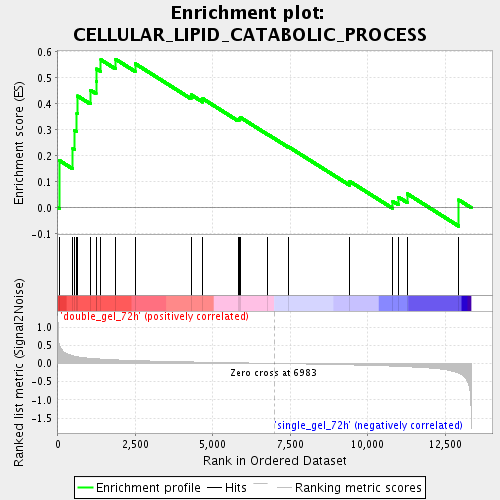
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_72h_versus_single_gel_72h.class.cls #double_gel_72h_versus_single_gel_72h_repos |
| Phenotype | class.cls#double_gel_72h_versus_single_gel_72h_repos |
| Upregulated in class | double_gel_72h |
| GeneSet | CELLULAR_LIPID_CATABOLIC_PROCESS |
| Enrichment Score (ES) | 0.5717224 |
| Normalized Enrichment Score (NES) | 1.6332262 |
| Nominal p-value | 0.028077753 |
| FDR q-value | 0.058566004 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | HACL1 | HACL1 Entrez, Source | 2-hydroxyacyl-CoA lyase 1 | 54 | 0.488 | 0.1815 | Yes |
| 2 | ACADVL | ACADVL Entrez, Source | acyl-Coenzyme A dehydrogenase, very long chain | 482 | 0.207 | 0.2281 | Yes |
| 3 | HAO2 | HAO2 Entrez, Source | hydroxyacid oxidase 2 (long chain) | 536 | 0.195 | 0.2982 | Yes |
| 4 | ALDH5A1 | ALDH5A1 Entrez, Source | aldehyde dehydrogenase 5 family, member A1 (succinate-semialdehyde dehydrogenase) | 612 | 0.184 | 0.3626 | Yes |
| 5 | ECHS1 | ECHS1 Entrez, Source | enoyl Coenzyme A hydratase, short chain, 1, mitochondrial | 633 | 0.182 | 0.4303 | Yes |
| 6 | CPT1A | CPT1A Entrez, Source | carnitine palmitoyltransferase 1A (liver) | 1046 | 0.140 | 0.4527 | Yes |
| 7 | SMPD3 | SMPD3 Entrez, Source | sphingomyelin phosphodiesterase 3, neutral membrane (neutral sphingomyelinase II) | 1232 | 0.128 | 0.4874 | Yes |
| 8 | PPARA | PPARA Entrez, Source | peroxisome proliferative activated receptor, alpha | 1252 | 0.127 | 0.5341 | Yes |
| 9 | PPARD | PPARD Entrez, Source | peroxisome proliferative activated receptor, delta | 1387 | 0.119 | 0.5694 | Yes |
| 10 | AKR1D1 | AKR1D1 Entrez, Source | aldo-keto reductase family 1, member D1 (delta 4-3-ketosteroid-5-beta-reductase) | 1859 | 0.099 | 0.5717 | Yes |
| 11 | CPT1B | CPT1B Entrez, Source | carnitine palmitoyltransferase 1B (muscle) | 2496 | 0.080 | 0.5543 | No |
| 12 | ACADM | ACADM Entrez, Source | acyl-Coenzyme A dehydrogenase, C-4 to C-12 straight chain | 4299 | 0.041 | 0.4348 | No |
| 13 | HADHB | HADHB Entrez, Source | hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (trifunctional protein), beta subunit | 4675 | 0.035 | 0.4198 | No |
| 14 | APOA5 | APOA5 Entrez, Source | apolipoprotein A-V | 5815 | 0.017 | 0.3408 | No |
| 15 | ECH1 | ECH1 Entrez, Source | enoyl Coenzyme A hydratase 1, peroxisomal | 5859 | 0.016 | 0.3438 | No |
| 16 | ACADS | ACADS Entrez, Source | acyl-Coenzyme A dehydrogenase, C-2 to C-3 short chain | 5886 | 0.016 | 0.3479 | No |
| 17 | PRDX6 | PRDX6 Entrez, Source | peroxiredoxin 6 | 6768 | 0.003 | 0.2829 | No |
| 18 | NEU3 | NEU3 Entrez, Source | sialidase 3 (membrane sialidase) | 7444 | -0.007 | 0.2347 | No |
| 19 | PPT1 | PPT1 Entrez, Source | palmitoyl-protein thioesterase 1 (ceroid-lipofuscinosis, neuronal 1, infantile) | 9419 | -0.040 | 0.1017 | No |
| 20 | CEL | CEL Entrez, Source | carboxyl ester lipase (bile salt-stimulated lipase) | 10808 | -0.073 | 0.0251 | No |
| 21 | YWHAH | YWHAH Entrez, Source | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide | 10999 | -0.079 | 0.0407 | No |
| 22 | STS | STS Entrez, Source | steroid sulfatase (microsomal), arylsulfatase C, isozyme S | 11277 | -0.089 | 0.0539 | No |
| 23 | LYPLA3 | LYPLA3 Entrez, Source | lysophospholipase 3 (lysosomal phospholipase A2) | 12939 | -0.266 | 0.0303 | No |