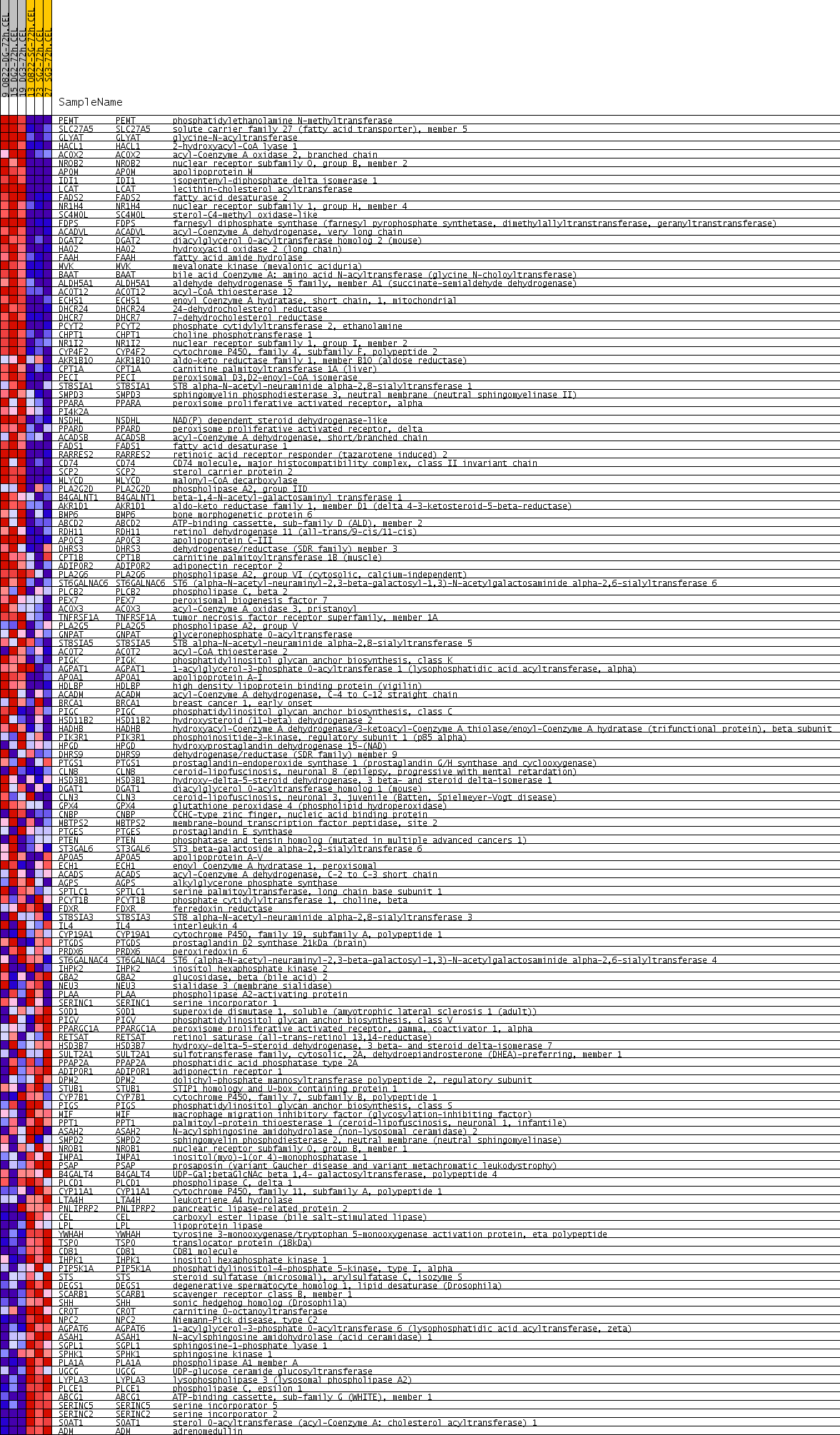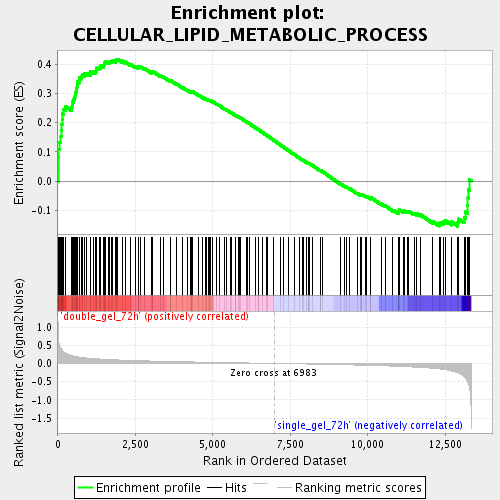
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_72h_versus_single_gel_72h.class.cls #double_gel_72h_versus_single_gel_72h_repos |
| Phenotype | class.cls#double_gel_72h_versus_single_gel_72h_repos |
| Upregulated in class | double_gel_72h |
| GeneSet | CELLULAR_LIPID_METABOLIC_PROCESS |
| Enrichment Score (ES) | 0.41687414 |
| Normalized Enrichment Score (NES) | 1.6985707 |
| Nominal p-value | 0.002967359 |
| FDR q-value | 0.03647462 |
| FWER p-Value | 0.999 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | PEMT | PEMT Entrez, Source | phosphatidylethanolamine N-methyltransferase | 8 | 0.798 | 0.0412 | Yes |
| 2 | SLC27A5 | SLC27A5 Entrez, Source | solute carrier family 27 (fatty acid transporter), member 5 | 9 | 0.779 | 0.0821 | Yes |
| 3 | GLYAT | GLYAT Entrez, Source | glycine-N-acyltransferase | 33 | 0.554 | 0.1094 | Yes |
| 4 | HACL1 | HACL1 Entrez, Source | 2-hydroxyacyl-CoA lyase 1 | 54 | 0.488 | 0.1335 | Yes |
| 5 | ACOX2 | ACOX2 Entrez, Source | acyl-Coenzyme A oxidase 2, branched chain | 95 | 0.419 | 0.1524 | Yes |
| 6 | NR0B2 | NR0B2 Entrez, Source | nuclear receptor subfamily 0, group B, member 2 | 104 | 0.416 | 0.1736 | Yes |
| 7 | APOM | APOM Entrez, Source | apolipoprotein M | 113 | 0.401 | 0.1940 | Yes |
| 8 | IDI1 | IDI1 Entrez, Source | isopentenyl-diphosphate delta isomerase 1 | 136 | 0.372 | 0.2119 | Yes |
| 9 | LCAT | LCAT Entrez, Source | lecithin-cholesterol acyltransferase | 142 | 0.365 | 0.2306 | Yes |
| 10 | FADS2 | FADS2 Entrez, Source | fatty acid desaturase 2 | 182 | 0.324 | 0.2447 | Yes |
| 11 | NR1H4 | NR1H4 Entrez, Source | nuclear receptor subfamily 1, group H, member 4 | 246 | 0.288 | 0.2550 | Yes |
| 12 | SC4MOL | SC4MOL Entrez, Source | sterol-C4-methyl oxidase-like | 444 | 0.218 | 0.2515 | Yes |
| 13 | FDPS | FDPS Entrez, Source | farnesyl diphosphate synthase (farnesyl pyrophosphate synthetase, dimethylallyltranstransferase, geranyltranstransferase) | 462 | 0.212 | 0.2613 | Yes |
| 14 | ACADVL | ACADVL Entrez, Source | acyl-Coenzyme A dehydrogenase, very long chain | 482 | 0.207 | 0.2707 | Yes |
| 15 | DGAT2 | DGAT2 Entrez, Source | diacylglycerol O-acyltransferase homolog 2 (mouse) | 503 | 0.201 | 0.2798 | Yes |
| 16 | HAO2 | HAO2 Entrez, Source | hydroxyacid oxidase 2 (long chain) | 536 | 0.195 | 0.2875 | Yes |
| 17 | FAAH | FAAH Entrez, Source | fatty acid amide hydrolase | 553 | 0.192 | 0.2964 | Yes |
| 18 | MVK | MVK Entrez, Source | mevalonate kinase (mevalonic aciduria) | 578 | 0.188 | 0.3044 | Yes |
| 19 | BAAT | BAAT Entrez, Source | bile acid Coenzyme A: amino acid N-acyltransferase (glycine N-choloyltransferase) | 597 | 0.186 | 0.3128 | Yes |
| 20 | ALDH5A1 | ALDH5A1 Entrez, Source | aldehyde dehydrogenase 5 family, member A1 (succinate-semialdehyde dehydrogenase) | 612 | 0.184 | 0.3215 | Yes |
| 21 | ACOT12 | ACOT12 Entrez, Source | acyl-CoA thioesterase 12 | 619 | 0.184 | 0.3306 | Yes |
| 22 | ECHS1 | ECHS1 Entrez, Source | enoyl Coenzyme A hydratase, short chain, 1, mitochondrial | 633 | 0.182 | 0.3392 | Yes |
| 23 | DHCR24 | DHCR24 Entrez, Source | 24-dehydrocholesterol reductase | 685 | 0.175 | 0.3445 | Yes |
| 24 | DHCR7 | DHCR7 Entrez, Source | 7-dehydrocholesterol reductase | 693 | 0.174 | 0.3531 | Yes |
| 25 | PCYT2 | PCYT2 Entrez, Source | phosphate cytidylyltransferase 2, ethanolamine | 748 | 0.168 | 0.3578 | Yes |
| 26 | CHPT1 | CHPT1 Entrez, Source | choline phosphotransferase 1 | 778 | 0.165 | 0.3642 | Yes |
| 27 | NR1I2 | NR1I2 Entrez, Source | nuclear receptor subfamily 1, group I, member 2 | 855 | 0.157 | 0.3667 | Yes |
| 28 | CYP4F2 | CYP4F2 Entrez, Source | cytochrome P450, family 4, subfamily F, polypeptide 2 | 933 | 0.150 | 0.3687 | Yes |
| 29 | AKR1B10 | AKR1B10 Entrez, Source | aldo-keto reductase family 1, member B10 (aldose reductase) | 1044 | 0.141 | 0.3678 | Yes |
| 30 | CPT1A | CPT1A Entrez, Source | carnitine palmitoyltransferase 1A (liver) | 1046 | 0.140 | 0.3751 | Yes |
| 31 | PECI | PECI Entrez, Source | peroxisomal D3,D2-enoyl-CoA isomerase | 1151 | 0.133 | 0.3742 | Yes |
| 32 | ST8SIA1 | ST8SIA1 Entrez, Source | ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1 | 1223 | 0.128 | 0.3755 | Yes |
| 33 | SMPD3 | SMPD3 Entrez, Source | sphingomyelin phosphodiesterase 3, neutral membrane (neutral sphingomyelinase II) | 1232 | 0.128 | 0.3816 | Yes |
| 34 | PPARA | PPARA Entrez, Source | peroxisome proliferative activated receptor, alpha | 1252 | 0.127 | 0.3868 | Yes |
| 35 | PI4K2A | 1329 | 0.123 | 0.3875 | Yes | ||
| 36 | NSDHL | NSDHL Entrez, Source | NAD(P) dependent steroid dehydrogenase-like | 1370 | 0.121 | 0.3907 | Yes |
| 37 | PPARD | PPARD Entrez, Source | peroxisome proliferative activated receptor, delta | 1387 | 0.119 | 0.3958 | Yes |
| 38 | ACADSB | ACADSB Entrez, Source | acyl-Coenzyme A dehydrogenase, short/branched chain | 1480 | 0.114 | 0.3948 | Yes |
| 39 | FADS1 | FADS1 Entrez, Source | fatty acid desaturase 1 | 1495 | 0.114 | 0.3997 | Yes |
| 40 | RARRES2 | RARRES2 Entrez, Source | retinoic acid receptor responder (tazarotene induced) 2 | 1501 | 0.113 | 0.4052 | Yes |
| 41 | CD74 | CD74 Entrez, Source | CD74 molecule, major histocompatibility complex, class II invariant chain | 1524 | 0.112 | 0.4094 | Yes |
| 42 | SCP2 | SCP2 Entrez, Source | sterol carrier protein 2 | 1632 | 0.107 | 0.4070 | Yes |
| 43 | MLYCD | MLYCD Entrez, Source | malonyl-CoA decarboxylase | 1678 | 0.106 | 0.4091 | Yes |
| 44 | PLA2G2D | PLA2G2D Entrez, Source | phospholipase A2, group IID | 1734 | 0.104 | 0.4104 | Yes |
| 45 | B4GALNT1 | B4GALNT1 Entrez, Source | beta-1,4-N-acetyl-galactosaminyl transferase 1 | 1769 | 0.102 | 0.4131 | Yes |
| 46 | AKR1D1 | AKR1D1 Entrez, Source | aldo-keto reductase family 1, member D1 (delta 4-3-ketosteroid-5-beta-reductase) | 1859 | 0.099 | 0.4116 | Yes |
| 47 | BMP6 | BMP6 Entrez, Source | bone morphogenetic protein 6 | 1877 | 0.098 | 0.4155 | Yes |
| 48 | ABCD2 | ABCD2 Entrez, Source | ATP-binding cassette, sub-family D (ALD), member 2 | 1926 | 0.096 | 0.4169 | Yes |
| 49 | RDH11 | RDH11 Entrez, Source | retinol dehydrogenase 11 (all-trans/9-cis/11-cis) | 2069 | 0.092 | 0.4109 | No |
| 50 | APOC3 | APOC3 Entrez, Source | apolipoprotein C-III | 2176 | 0.089 | 0.4075 | No |
| 51 | DHRS3 | DHRS3 Entrez, Source | dehydrogenase/reductase (SDR family) member 3 | 2353 | 0.084 | 0.3986 | No |
| 52 | CPT1B | CPT1B Entrez, Source | carnitine palmitoyltransferase 1B (muscle) | 2496 | 0.080 | 0.3920 | No |
| 53 | ADIPOR2 | ADIPOR2 Entrez, Source | adiponectin receptor 2 | 2598 | 0.076 | 0.3884 | No |
| 54 | PLA2G6 | PLA2G6 Entrez, Source | phospholipase A2, group VI (cytosolic, calcium-independent) | 2615 | 0.076 | 0.3911 | No |
| 55 | ST6GALNAC6 | ST6GALNAC6 Entrez, Source | ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 | 2672 | 0.074 | 0.3908 | No |
| 56 | PLCB2 | PLCB2 Entrez, Source | phospholipase C, beta 2 | 2782 | 0.072 | 0.3863 | No |
| 57 | PEX7 | PEX7 Entrez, Source | peroxisomal biogenesis factor 7 | 3029 | 0.066 | 0.3711 | No |
| 58 | ACOX3 | ACOX3 Entrez, Source | acyl-Coenzyme A oxidase 3, pristanoyl | 3046 | 0.066 | 0.3733 | No |
| 59 | TNFRSF1A | TNFRSF1A Entrez, Source | tumor necrosis factor receptor superfamily, member 1A | 3063 | 0.065 | 0.3755 | No |
| 60 | PLA2G5 | PLA2G5 Entrez, Source | phospholipase A2, group V | 3301 | 0.060 | 0.3607 | No |
| 61 | GNPAT | GNPAT Entrez, Source | glyceronephosphate O-acyltransferase | 3398 | 0.058 | 0.3564 | No |
| 62 | ST8SIA5 | ST8SIA5 Entrez, Source | ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 5 | 3625 | 0.053 | 0.3421 | No |
| 63 | ACOT2 | ACOT2 Entrez, Source | acyl-CoA thioesterase 2 | 3642 | 0.053 | 0.3437 | No |
| 64 | PIGK | PIGK Entrez, Source | phosphatidylinositol glycan anchor biosynthesis, class K | 3814 | 0.050 | 0.3333 | No |
| 65 | AGPAT1 | AGPAT1 Entrez, Source | 1-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha) | 4028 | 0.046 | 0.3195 | No |
| 66 | APOA1 | APOA1 Entrez, Source | apolipoprotein A-I | 4169 | 0.043 | 0.3112 | No |
| 67 | HDLBP | HDLBP Entrez, Source | high density lipoprotein binding protein (vigilin) | 4270 | 0.042 | 0.3058 | No |
| 68 | ACADM | ACADM Entrez, Source | acyl-Coenzyme A dehydrogenase, C-4 to C-12 straight chain | 4299 | 0.041 | 0.3059 | No |
| 69 | BRCA1 | BRCA1 Entrez, Source | breast cancer 1, early onset | 4326 | 0.041 | 0.3060 | No |
| 70 | PIGC | PIGC Entrez, Source | phosphatidylinositol glycan anchor biosynthesis, class C | 4338 | 0.040 | 0.3073 | No |
| 71 | HSD11B2 | HSD11B2 Entrez, Source | hydroxysteroid (11-beta) dehydrogenase 2 | 4542 | 0.037 | 0.2939 | No |
| 72 | HADHB | HADHB Entrez, Source | hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (trifunctional protein), beta subunit | 4675 | 0.035 | 0.2857 | No |
| 73 | PIK3R1 | PIK3R1 Entrez, Source | phosphoinositide-3-kinase, regulatory subunit 1 (p85 alpha) | 4760 | 0.033 | 0.2810 | No |
| 74 | HPGD | HPGD Entrez, Source | hydroxyprostaglandin dehydrogenase 15-(NAD) | 4803 | 0.032 | 0.2796 | No |
| 75 | DHRS9 | DHRS9 Entrez, Source | dehydrogenase/reductase (SDR family) member 9 | 4865 | 0.031 | 0.2766 | No |
| 76 | PTGS1 | PTGS1 Entrez, Source | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | 4887 | 0.031 | 0.2766 | No |
| 77 | CLN8 | CLN8 Entrez, Source | ceroid-lipofuscinosis, neuronal 8 (epilepsy, progressive with mental retardation) | 4936 | 0.030 | 0.2746 | No |
| 78 | HSD3B1 | HSD3B1 Entrez, Source | hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 1 | 4997 | 0.029 | 0.2715 | No |
| 79 | DGAT1 | DGAT1 Entrez, Source | diacylglycerol O-acyltransferase homolog 1 (mouse) | 5118 | 0.027 | 0.2639 | No |
| 80 | CLN3 | CLN3 Entrez, Source | ceroid-lipofuscinosis, neuronal 3, juvenile (Batten, Spielmeyer-Vogt disease) | 5208 | 0.026 | 0.2585 | No |
| 81 | GPX4 | GPX4 Entrez, Source | glutathione peroxidase 4 (phospholipid hydroperoxidase) | 5373 | 0.024 | 0.2473 | No |
| 82 | CNBP | CNBP Entrez, Source | CCHC-type zinc finger, nucleic acid binding protein | 5438 | 0.023 | 0.2436 | No |
| 83 | MBTPS2 | MBTPS2 Entrez, Source | membrane-bound transcription factor peptidase, site 2 | 5566 | 0.021 | 0.2351 | No |
| 84 | PTGES | PTGES Entrez, Source | prostaglandin E synthase | 5602 | 0.020 | 0.2335 | No |
| 85 | PTEN | PTEN Entrez, Source | phosphatase and tensin homolog (mutated in multiple advanced cancers 1) | 5720 | 0.018 | 0.2256 | No |
| 86 | ST3GAL6 | ST3GAL6 Entrez, Source | ST3 beta-galactoside alpha-2,3-sialyltransferase 6 | 5745 | 0.018 | 0.2247 | No |
| 87 | APOA5 | APOA5 Entrez, Source | apolipoprotein A-V | 5815 | 0.017 | 0.2204 | No |
| 88 | ECH1 | ECH1 Entrez, Source | enoyl Coenzyme A hydratase 1, peroxisomal | 5859 | 0.016 | 0.2180 | No |
| 89 | ACADS | ACADS Entrez, Source | acyl-Coenzyme A dehydrogenase, C-2 to C-3 short chain | 5886 | 0.016 | 0.2169 | No |
| 90 | AGPS | AGPS Entrez, Source | alkylglycerone phosphate synthase | 6079 | 0.013 | 0.2030 | No |
| 91 | SPTLC1 | SPTLC1 Entrez, Source | serine palmitoyltransferase, long chain base subunit 1 | 6112 | 0.013 | 0.2012 | No |
| 92 | PCYT1B | PCYT1B Entrez, Source | phosphate cytidylyltransferase 1, choline, beta | 6186 | 0.012 | 0.1963 | No |
| 93 | FDXR | FDXR Entrez, Source | ferredoxin reductase | 6391 | 0.008 | 0.1812 | No |
| 94 | ST8SIA3 | ST8SIA3 Entrez, Source | ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 | 6457 | 0.007 | 0.1767 | No |
| 95 | IL4 | IL4 Entrez, Source | interleukin 4 | 6466 | 0.007 | 0.1764 | No |
| 96 | CYP19A1 | CYP19A1 Entrez, Source | cytochrome P450, family 19, subfamily A, polypeptide 1 | 6596 | 0.005 | 0.1669 | No |
| 97 | PTGDS | PTGDS Entrez, Source | prostaglandin D2 synthase 21kDa (brain) | 6738 | 0.003 | 0.1564 | No |
| 98 | PRDX6 | PRDX6 Entrez, Source | peroxiredoxin 6 | 6768 | 0.003 | 0.1544 | No |
| 99 | ST6GALNAC4 | ST6GALNAC4 Entrez, Source | ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 4 | 6968 | 0.000 | 0.1393 | No |
| 100 | IHPK2 | IHPK2 Entrez, Source | inositol hexaphosphate kinase 2 | 7190 | -0.003 | 0.1227 | No |
| 101 | GBA2 | GBA2 Entrez, Source | glucosidase, beta (bile acid) 2 | 7269 | -0.004 | 0.1170 | No |
| 102 | NEU3 | NEU3 Entrez, Source | sialidase 3 (membrane sialidase) | 7444 | -0.007 | 0.1042 | No |
| 103 | PLAA | PLAA Entrez, Source | phospholipase A2-activating protein | 7627 | -0.009 | 0.0909 | No |
| 104 | SERINC1 | SERINC1 Entrez, Source | serine incorporator 1 | 7788 | -0.012 | 0.0793 | No |
| 105 | SOD1 | SOD1 Entrez, Source | superoxide dismutase 1, soluble (amyotrophic lateral sclerosis 1 (adult)) | 7883 | -0.013 | 0.0729 | No |
| 106 | PIGV | PIGV Entrez, Source | phosphatidylinositol glycan anchor biosynthesis, class V | 7912 | -0.014 | 0.0715 | No |
| 107 | PPARGC1A | PPARGC1A Entrez, Source | peroxisome proliferative activated receptor, gamma, coactivator 1, alpha | 8036 | -0.015 | 0.0630 | No |
| 108 | RETSAT | RETSAT Entrez, Source | retinol saturase (all-trans-retinol 13,14-reductase) | 8072 | -0.016 | 0.0612 | No |
| 109 | HSD3B7 | HSD3B7 Entrez, Source | hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 | 8089 | -0.016 | 0.0608 | No |
| 110 | SULT2A1 | SULT2A1 Entrez, Source | sulfotransferase family, cytosolic, 2A, dehydroepiandrosterone (DHEA)-preferring, member 1 | 8105 | -0.016 | 0.0605 | No |
| 111 | PPAP2A | PPAP2A Entrez, Source | phosphatidic acid phosphatase type 2A | 8226 | -0.018 | 0.0524 | No |
| 112 | ADIPOR1 | ADIPOR1 Entrez, Source | adiponectin receptor 1 | 8460 | -0.022 | 0.0359 | No |
| 113 | DPM2 | DPM2 Entrez, Source | dolichyl-phosphate mannosyltransferase polypeptide 2, regulatory subunit | 8478 | -0.023 | 0.0358 | No |
| 114 | STUB1 | STUB1 Entrez, Source | STIP1 homology and U-box containing protein 1 | 8548 | -0.024 | 0.0318 | No |
| 115 | CYP7B1 | CYP7B1 Entrez, Source | cytochrome P450, family 7, subfamily B, polypeptide 1 | 9114 | -0.034 | -0.0092 | No |
| 116 | PIGS | PIGS Entrez, Source | phosphatidylinositol glycan anchor biosynthesis, class S | 9263 | -0.037 | -0.0185 | No |
| 117 | MIF | MIF Entrez, Source | macrophage migration inhibitory factor (glycosylation-inhibiting factor) | 9316 | -0.038 | -0.0205 | No |
| 118 | PPT1 | PPT1 Entrez, Source | palmitoyl-protein thioesterase 1 (ceroid-lipofuscinosis, neuronal 1, infantile) | 9419 | -0.040 | -0.0261 | No |
| 119 | ASAH2 | ASAH2 Entrez, Source | N-acylsphingosine amidohydrolase (non-lysosomal ceramidase) 2 | 9653 | -0.045 | -0.0414 | No |
| 120 | SMPD2 | SMPD2 Entrez, Source | sphingomyelin phosphodiesterase 2, neutral membrane (neutral sphingomyelinase) | 9756 | -0.047 | -0.0467 | No |
| 121 | NR0B1 | NR0B1 Entrez, Source | nuclear receptor subfamily 0, group B, member 1 | 9783 | -0.047 | -0.0462 | No |
| 122 | IMPA1 | IMPA1 Entrez, Source | inositol(myo)-1(or 4)-monophosphatase 1 | 9804 | -0.048 | -0.0452 | No |
| 123 | PSAP | PSAP Entrez, Source | prosaposin (variant Gaucher disease and variant metachromatic leukodystrophy) | 9922 | -0.051 | -0.0514 | No |
| 124 | B4GALT4 | B4GALT4 Entrez, Source | UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 4 | 9970 | -0.052 | -0.0523 | No |
| 125 | PLCD1 | PLCD1 Entrez, Source | phospholipase C, delta 1 | 10095 | -0.054 | -0.0588 | No |
| 126 | CYP11A1 | CYP11A1 Entrez, Source | cytochrome P450, family 11, subfamily A, polypeptide 1 | 10102 | -0.054 | -0.0564 | No |
| 127 | LTA4H | LTA4H Entrez, Source | leukotriene A4 hydrolase | 10430 | -0.062 | -0.0780 | No |
| 128 | PNLIPRP2 | PNLIPRP2 Entrez, Source | pancreatic lipase-related protein 2 | 10564 | -0.066 | -0.0846 | No |
| 129 | CEL | CEL Entrez, Source | carboxyl ester lipase (bile salt-stimulated lipase) | 10808 | -0.073 | -0.0992 | No |
| 130 | LPL | LPL Entrez, Source | lipoprotein lipase | 10988 | -0.078 | -0.1087 | No |
| 131 | YWHAH | YWHAH Entrez, Source | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide | 10999 | -0.079 | -0.1053 | No |
| 132 | TSPO | TSPO Entrez, Source | translocator protein (18kDa) | 11004 | -0.079 | -0.1015 | No |
| 133 | CD81 | CD81 Entrez, Source | CD81 molecule | 11022 | -0.079 | -0.0986 | No |
| 134 | IHPK1 | IHPK1 Entrez, Source | inositol hexaphosphate kinase 1 | 11145 | -0.083 | -0.1035 | No |
| 135 | PIP5K1A | PIP5K1A Entrez, Source | phosphatidylinositol-4-phosphate 5-kinase, type I, alpha | 11170 | -0.084 | -0.1009 | No |
| 136 | STS | STS Entrez, Source | steroid sulfatase (microsomal), arylsulfatase C, isozyme S | 11277 | -0.089 | -0.1042 | No |
| 137 | DEGS1 | DEGS1 Entrez, Source | degenerative spermatocyte homolog 1, lipid desaturase (Drosophila) | 11324 | -0.092 | -0.1029 | No |
| 138 | SCARB1 | SCARB1 Entrez, Source | scavenger receptor class B, member 1 | 11498 | -0.099 | -0.1108 | No |
| 139 | SHH | SHH Entrez, Source | sonic hedgehog homolog (Drosophila) | 11581 | -0.103 | -0.1116 | No |
| 140 | CROT | CROT Entrez, Source | carnitine O-octanoyltransferase | 11691 | -0.109 | -0.1142 | No |
| 141 | NPC2 | NPC2 Entrez, Source | Niemann-Pick disease, type C2 | 12096 | -0.131 | -0.1380 | No |
| 142 | AGPAT6 | AGPAT6 Entrez, Source | 1-acylglycerol-3-phosphate O-acyltransferase 6 (lysophosphatidic acid acyltransferase, zeta) | 12308 | -0.149 | -0.1461 | No |
| 143 | ASAH1 | ASAH1 Entrez, Source | N-acylsphingosine amidohydrolase (acid ceramidase) 1 | 12353 | -0.153 | -0.1414 | No |
| 144 | SGPL1 | SGPL1 Entrez, Source | sphingosine-1-phosphate lyase 1 | 12434 | -0.162 | -0.1390 | No |
| 145 | SPHK1 | SPHK1 Entrez, Source | sphingosine kinase 1 | 12507 | -0.172 | -0.1355 | No |
| 146 | PLA1A | PLA1A Entrez, Source | phospholipase A1 member A | 12708 | -0.206 | -0.1398 | No |
| 147 | UGCG | UGCG Entrez, Source | UDP-glucose ceramide glucosyltransferase | 12906 | -0.254 | -0.1414 | No |
| 148 | LYPLA3 | LYPLA3 Entrez, Source | lysophospholipase 3 (lysosomal phospholipase A2) | 12939 | -0.266 | -0.1299 | No |
| 149 | PLCE1 | PLCE1 Entrez, Source | phospholipase C, epsilon 1 | 13110 | -0.363 | -0.1237 | No |
| 150 | ABCG1 | ABCG1 Entrez, Source | ATP-binding cassette, sub-family G (WHITE), member 1 | 13162 | -0.417 | -0.1057 | No |
| 151 | SERINC5 | SERINC5 Entrez, Source | serine incorporator 5 | 13225 | -0.503 | -0.0840 | No |
| 152 | SERINC2 | SERINC2 Entrez, Source | serine incorporator 2 | 13234 | -0.520 | -0.0574 | No |
| 153 | SOAT1 | SOAT1 Entrez, Source | sterol O-acyltransferase (acyl-Coenzyme A: cholesterol acyltransferase) 1 | 13248 | -0.551 | -0.0295 | No |
| 154 | ADM | ADM Entrez, Source | adrenomedullin | 13287 | -0.697 | 0.0042 | No |