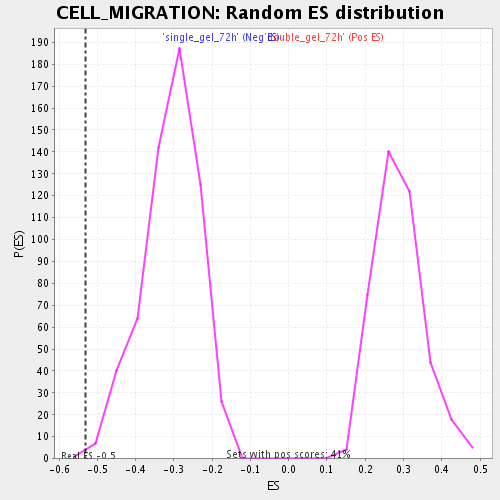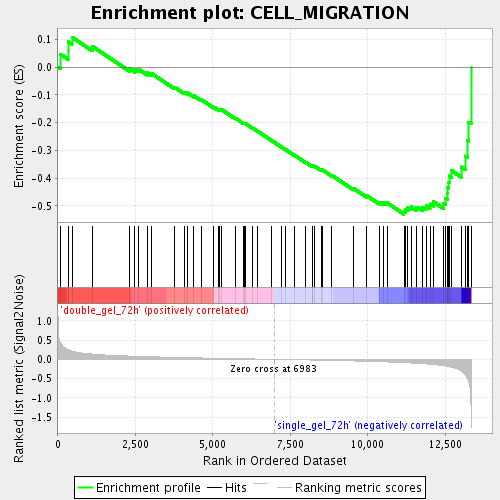
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_72h_versus_single_gel_72h.class.cls #double_gel_72h_versus_single_gel_72h_repos |
| Phenotype | class.cls#double_gel_72h_versus_single_gel_72h_repos |
| Upregulated in class | single_gel_72h |
| GeneSet | CELL_MIGRATION |
| Enrichment Score (ES) | -0.5299964 |
| Normalized Enrichment Score (NES) | -1.7204789 |
| Nominal p-value | 0.0016891892 |
| FDR q-value | 0.055886727 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | PLG | PLG Entrez, Source | plasminogen | 81 | 0.434 | 0.0468 | No |
| 2 | HMGCR | HMGCR Entrez, Source | 3-hydroxy-3-methylglutaryl-Coenzyme A reductase | 328 | 0.257 | 0.0597 | No |
| 3 | PPAP2B | PPAP2B Entrez, Source | phosphatidic acid phosphatase type 2B | 329 | 0.257 | 0.0911 | No |
| 4 | CDK5R1 | CDK5R1 Entrez, Source | cyclin-dependent kinase 5, regulatory subunit 1 (p35) | 455 | 0.214 | 0.1078 | No |
| 5 | PF4 | PF4 Entrez, Source | platelet factor 4 (chemokine (C-X-C motif) ligand 4) | 1114 | 0.136 | 0.0748 | No |
| 6 | TBX5 | TBX5 Entrez, Source | T-box 5 | 2299 | 0.085 | -0.0039 | No |
| 7 | GTPBP4 | GTPBP4 Entrez, Source | GTP binding protein 4 | 2484 | 0.080 | -0.0080 | No |
| 8 | CNTN4 | CNTN4 Entrez, Source | contactin 4 | 2609 | 0.076 | -0.0081 | No |
| 9 | SFTPD | SFTPD Entrez, Source | surfactant, pulmonary-associated protein D | 2883 | 0.069 | -0.0202 | No |
| 10 | CALCA | CALCA Entrez, Source | calcitonin/calcitonin-related polypeptide, alpha | 3026 | 0.066 | -0.0228 | No |
| 11 | SLIT2 | SLIT2 Entrez, Source | slit homolog 2 (Drosophila) | 3770 | 0.050 | -0.0726 | No |
| 12 | EGFR | EGFR Entrez, Source | epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | 4079 | 0.045 | -0.0904 | No |
| 13 | ACVRL1 | ACVRL1 Entrez, Source | activin A receptor type II-like 1 | 4179 | 0.043 | -0.0925 | No |
| 14 | IL10 | IL10 Entrez, Source | interleukin 10 | 4386 | 0.040 | -0.1032 | No |
| 15 | UNC5C | UNC5C Entrez, Source | unc-5 homolog C (C. elegans) | 4638 | 0.035 | -0.1178 | No |
| 16 | SYK | SYK Entrez, Source | spleen tyrosine kinase | 5035 | 0.029 | -0.1441 | No |
| 17 | NRXN3 | NRXN3 Entrez, Source | neurexin 3 | 5181 | 0.027 | -0.1518 | No |
| 18 | GDNF | GDNF Entrez, Source | glial cell derived neurotrophic factor | 5229 | 0.026 | -0.1522 | No |
| 19 | IL12A | IL12A Entrez, Source | interleukin 12A (natural killer cell stimulatory factor 1, cytotoxic lymphocyte maturation factor 1, p35) | 5271 | 0.025 | -0.1522 | No |
| 20 | PTEN | PTEN Entrez, Source | phosphatase and tensin homolog (mutated in multiple advanced cancers 1) | 5720 | 0.018 | -0.1837 | No |
| 21 | IL12B | IL12B Entrez, Source | interleukin 12B (natural killer cell stimulatory factor 2, cytotoxic lymphocyte maturation factor 2, p40) | 6003 | 0.014 | -0.2032 | No |
| 22 | NF2 | NF2 Entrez, Source | neurofibromin 2 (bilateral acoustic neuroma) | 6018 | 0.014 | -0.2026 | No |
| 23 | SCYE1 | SCYE1 Entrez, Source | small inducible cytokine subfamily E, member 1 (endothelial monocyte-activating) | 6040 | 0.014 | -0.2025 | No |
| 24 | DPYSL5 | DPYSL5 Entrez, Source | dihydropyrimidinase-like 5 | 6290 | 0.010 | -0.2201 | No |
| 25 | NRD1 | NRD1 Entrez, Source | nardilysin (N-arginine dibasic convertase) | 6449 | 0.007 | -0.2311 | No |
| 26 | THBS4 | THBS4 Entrez, Source | thrombospondin 4 | 6884 | 0.002 | -0.2636 | No |
| 27 | NRXN1 | NRXN1 Entrez, Source | neurexin 1 | 7204 | -0.003 | -0.2872 | No |
| 28 | NRTN | NRTN Entrez, Source | neurturin | 7360 | -0.005 | -0.2982 | No |
| 29 | FEZ1 | FEZ1 Entrez, Source | fasciculation and elongation protein zeta 1 (zygin I) | 7623 | -0.009 | -0.3168 | No |
| 30 | NRP2 | NRP2 Entrez, Source | neuropilin 2 | 8004 | -0.015 | -0.3436 | No |
| 31 | ENPEP | ENPEP Entrez, Source | glutamyl aminopeptidase (aminopeptidase A) | 8208 | -0.018 | -0.3566 | No |
| 32 | PPAP2A | PPAP2A Entrez, Source | phosphatidic acid phosphatase type 2A | 8226 | -0.018 | -0.3557 | No |
| 33 | ITGB2 | ITGB2 Entrez, Source | integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) | 8281 | -0.020 | -0.3573 | No |
| 34 | TNFSF12 | TNFSF12 Entrez, Source | tumor necrosis factor (ligand) superfamily, member 12 | 8504 | -0.023 | -0.3712 | No |
| 35 | ABI3 | ABI3 Entrez, Source | ABI gene family, member 3 | 8535 | -0.024 | -0.3706 | No |
| 36 | UBB | UBB Entrez, Source | ubiquitin B | 8839 | -0.029 | -0.3899 | No |
| 37 | SCG2 | SCG2 Entrez, Source | secretogranin II (chromogranin C) | 9523 | -0.042 | -0.4362 | No |
| 38 | FEZ2 | FEZ2 Entrez, Source | fasciculation and elongation protein zeta 2 (zygin II) | 9964 | -0.051 | -0.4631 | No |
| 39 | IL8RB | IL8RB Entrez, Source | interleukin 8 receptor, beta | 10379 | -0.061 | -0.4868 | No |
| 40 | SLIT1 | SLIT1 Entrez, Source | slit homolog 1 (Drosophila) | 10513 | -0.064 | -0.4890 | No |
| 41 | SPON2 | SPON2 Entrez, Source | spondin 2, extracellular matrix protein | 10621 | -0.067 | -0.4888 | No |
| 42 | THY1 | THY1 Entrez, Source | Thy-1 cell surface antigen | 11169 | -0.084 | -0.5197 | Yes |
| 43 | NRP1 | NRP1 Entrez, Source | neuropilin 1 | 11218 | -0.087 | -0.5128 | Yes |
| 44 | SEMA4F | SEMA4F Entrez, Source | sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4F | 11276 | -0.089 | -0.5062 | Yes |
| 45 | ITGB1 | ITGB1 Entrez, Source | integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) | 11396 | -0.095 | -0.5036 | Yes |
| 46 | SHH | SHH Entrez, Source | sonic hedgehog homolog (Drosophila) | 11581 | -0.103 | -0.5048 | Yes |
| 47 | OTX2 | OTX2 Entrez, Source | orthodenticle homolog 2 (Drosophila) | 11767 | -0.112 | -0.5051 | Yes |
| 48 | NF1 | NF1 Entrez, Source | neurofibromin 1 (neurofibromatosis, von Recklinghausen disease, Watson disease) | 11882 | -0.118 | -0.4993 | Yes |
| 49 | BCAR1 | BCAR1 Entrez, Source | breast cancer anti-estrogen resistance 1 | 12011 | -0.126 | -0.4935 | Yes |
| 50 | TGFB2 | TGFB2 Entrez, Source | transforming growth factor, beta 2 | 12112 | -0.132 | -0.4850 | Yes |
| 51 | MYH9 | MYH9 Entrez, Source | myosin, heavy chain 9, non-muscle | 12456 | -0.165 | -0.4907 | Yes |
| 52 | SPHK1 | SPHK1 Entrez, Source | sphingosine kinase 1 | 12507 | -0.172 | -0.4735 | Yes |
| 53 | CX3CL1 | CX3CL1 Entrez, Source | chemokine (C-X3-C motif) ligand 1 | 12570 | -0.180 | -0.4562 | Yes |
| 54 | CLIC4 | CLIC4 Entrez, Source | chloride intracellular channel 4 | 12577 | -0.181 | -0.4346 | Yes |
| 55 | CKLF | CKLF Entrez, Source | chemokine-like factor | 12616 | -0.187 | -0.4146 | Yes |
| 56 | CDH13 | CDH13 Entrez, Source | cadherin 13, H-cadherin (heart) | 12630 | -0.190 | -0.3925 | Yes |
| 57 | VEGFC | VEGFC Entrez, Source | vascular endothelial growth factor C | 12697 | -0.204 | -0.3725 | Yes |
| 58 | LAMC1 | LAMC1 Entrez, Source | laminin, gamma 1 (formerly LAMB2) | 13018 | -0.300 | -0.3601 | Yes |
| 59 | ABI2 | ABI2 Entrez, Source | abl interactor 2 | 13159 | -0.411 | -0.3205 | Yes |
| 60 | RTN4 | RTN4 Entrez, Source | reticulon 4 | 13219 | -0.495 | -0.2645 | Yes |
| 61 | NEXN | NEXN Entrez, Source | nexilin (F actin binding protein) | 13256 | -0.568 | -0.1979 | Yes |
| 62 | CD24 | CD24 Entrez, Source | CD24 molecule | 13341 | -1.675 | 0.0001 | Yes |