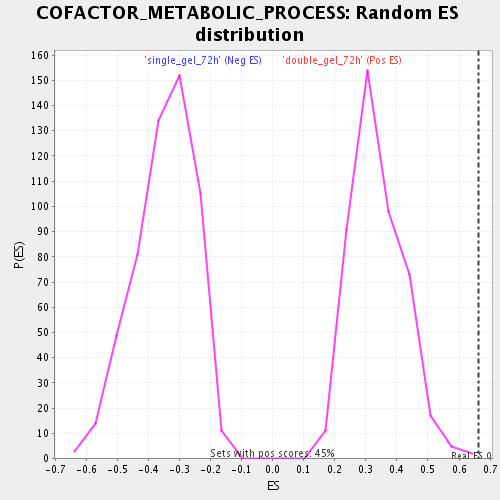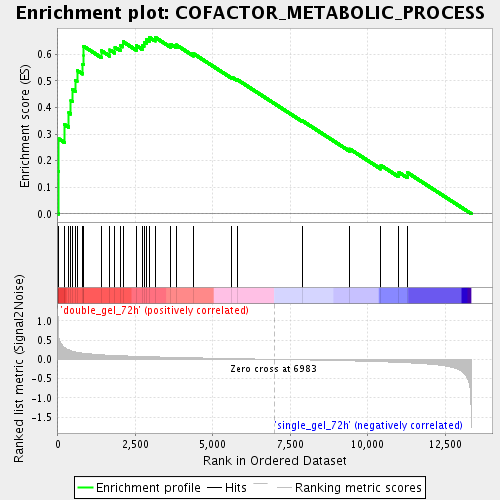
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_72h_versus_single_gel_72h.class.cls #double_gel_72h_versus_single_gel_72h_repos |
| Phenotype | class.cls#double_gel_72h_versus_single_gel_72h_repos |
| Upregulated in class | double_gel_72h |
| GeneSet | COFACTOR_METABOLIC_PROCESS |
| Enrichment Score (ES) | 0.66375804 |
| Normalized Enrichment Score (NES) | 1.9679871 |
| Nominal p-value | 0.0022172949 |
| FDR q-value | 0.0040315245 |
| FWER p-Value | 0.204 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | FTCD | FTCD Entrez, Source | formiminotransferase cyclodeaminase | 15 | 0.731 | 0.1604 | Yes |
| 2 | GLYAT | GLYAT Entrez, Source | glycine-N-acyltransferase | 33 | 0.554 | 0.2815 | Yes |
| 3 | ACLY | ACLY Entrez, Source | ATP citrate lyase | 212 | 0.305 | 0.3354 | Yes |
| 4 | MOCS2 | MOCS2 Entrez, Source | molybdenum cofactor synthesis 2 | 352 | 0.248 | 0.3798 | Yes |
| 5 | ALAS1 | ALAS1 Entrez, Source | aminolevulinate, delta-, synthase 1 | 419 | 0.224 | 0.4244 | Yes |
| 6 | ALDH1L1 | ALDH1L1 Entrez, Source | aldehyde dehydrogenase 1 family, member L1 | 465 | 0.211 | 0.4677 | Yes |
| 7 | PDHB | PDHB Entrez, Source | pyruvate dehydrogenase (lipoamide) beta | 563 | 0.190 | 0.5024 | Yes |
| 8 | ACOT12 | ACOT12 Entrez, Source | acyl-CoA thioesterase 12 | 619 | 0.184 | 0.5389 | Yes |
| 9 | ME1 | ME1 Entrez, Source | malic enzyme 1, NADP(+)-dependent, cytosolic | 800 | 0.163 | 0.5614 | Yes |
| 10 | GCLC | GCLC Entrez, Source | glutamate-cysteine ligase, catalytic subunit | 823 | 0.160 | 0.5951 | Yes |
| 11 | ALAD | ALAD Entrez, Source | aminolevulinate, delta-, dehydratase | 826 | 0.160 | 0.6303 | Yes |
| 12 | COQ3 | COQ3 Entrez, Source | coenzyme Q3 homolog, methyltransferase (S. cerevisiae) | 1406 | 0.118 | 0.6129 | Yes |
| 13 | MLYCD | MLYCD Entrez, Source | malonyl-CoA decarboxylase | 1678 | 0.106 | 0.6159 | Yes |
| 14 | GCLM | GCLM Entrez, Source | glutamate-cysteine ligase, modifier subunit | 1840 | 0.100 | 0.6258 | Yes |
| 15 | COQ7 | COQ7 Entrez, Source | coenzyme Q7 homolog, ubiquinone (yeast) | 2016 | 0.093 | 0.6333 | Yes |
| 16 | NFS1 | NFS1 Entrez, Source | NFS1 nitrogen fixation 1 (S. cerevisiae) | 2100 | 0.091 | 0.6471 | Yes |
| 17 | SDHD | SDHD Entrez, Source | succinate dehydrogenase complex, subunit D, integral membrane protein | 2529 | 0.079 | 0.6324 | Yes |
| 18 | GLRX2 | GLRX2 Entrez, Source | glutaredoxin 2 | 2722 | 0.073 | 0.6341 | Yes |
| 19 | SDHA | SDHA Entrez, Source | succinate dehydrogenase complex, subunit A, flavoprotein (Fp) | 2780 | 0.072 | 0.6457 | Yes |
| 20 | COASY | COASY Entrez, Source | Coenzyme A synthase | 2861 | 0.070 | 0.6551 | Yes |
| 21 | CPOX | CPOX Entrez, Source | coproporphyrinogen oxidase | 2946 | 0.068 | 0.6638 | Yes |
| 22 | ACO2 | ACO2 Entrez, Source | aconitase 2, mitochondrial | 3136 | 0.064 | 0.6636 | No |
| 23 | ACOT2 | ACOT2 Entrez, Source | acyl-CoA thioesterase 2 | 3642 | 0.053 | 0.6374 | No |
| 24 | UROS | UROS Entrez, Source | uroporphyrinogen III synthase (congenital erythropoietic porphyria) | 3820 | 0.049 | 0.6350 | No |
| 25 | BLVRA | BLVRA Entrez, Source | biliverdin reductase A | 4380 | 0.040 | 0.6018 | No |
| 26 | ALAS2 | ALAS2 Entrez, Source | aminolevulinate, delta-, synthase 2 (sideroblastic/hypochromic anemia) | 5606 | 0.020 | 0.5142 | No |
| 27 | SDHC | SDHC Entrez, Source | succinate dehydrogenase complex, subunit C, integral membrane protein, 15kDa | 5789 | 0.018 | 0.5044 | No |
| 28 | SOD1 | SOD1 Entrez, Source | superoxide dismutase 1, soluble (amyotrophic lateral sclerosis 1 (adult)) | 7883 | -0.013 | 0.3501 | No |
| 29 | PPT1 | PPT1 Entrez, Source | palmitoyl-protein thioesterase 1 (ceroid-lipofuscinosis, neuronal 1, infantile) | 9419 | -0.040 | 0.2437 | No |
| 30 | COX15 | COX15 Entrez, Source | COX15 homolog, cytochrome c oxidase assembly protein (yeast) | 10424 | -0.062 | 0.1819 | No |
| 31 | TSPO | TSPO Entrez, Source | translocator protein (18kDa) | 11004 | -0.079 | 0.1558 | No |
| 32 | GPX1 | GPX1 Entrez, Source | glutathione peroxidase 1 | 11279 | -0.089 | 0.1550 | No |