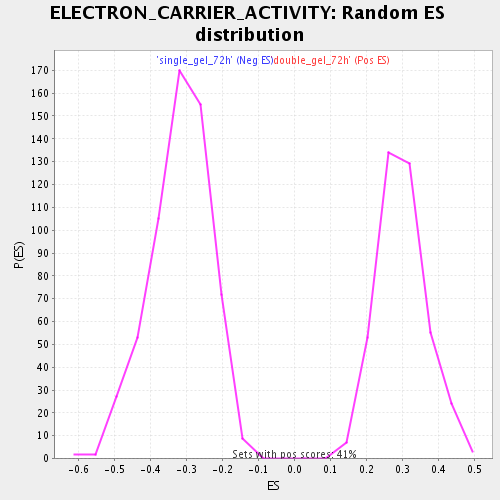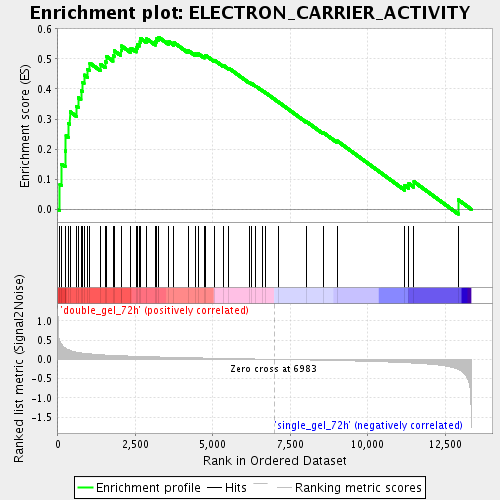
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_72h_versus_single_gel_72h.class.cls #double_gel_72h_versus_single_gel_72h_repos |
| Phenotype | class.cls#double_gel_72h_versus_single_gel_72h_repos |
| Upregulated in class | double_gel_72h |
| GeneSet | ELECTRON_CARRIER_ACTIVITY |
| Enrichment Score (ES) | 0.57188594 |
| Normalized Enrichment Score (NES) | 1.9177777 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0060897064 |
| FWER p-Value | 0.378 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | KMO | KMO Entrez, Source | kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) | 64 | 0.478 | 0.0826 | Yes |
| 2 | HAAO | HAAO Entrez, Source | 3-hydroxyanthranilate 3,4-dioxygenase | 120 | 0.390 | 0.1499 | Yes |
| 3 | SARDH | SARDH Entrez, Source | sarcosine dehydrogenase | 237 | 0.291 | 0.1943 | Yes |
| 4 | AKR7A2 | AKR7A2 Entrez, Source | aldo-keto reductase family 7, member A2 (aflatoxin aldehyde reductase) | 260 | 0.283 | 0.2445 | Yes |
| 5 | DMGDH | DMGDH Entrez, Source | dimethylglycine dehydrogenase | 334 | 0.255 | 0.2856 | Yes |
| 6 | CYP1A2 | CYP1A2 Entrez, Source | cytochrome P450, family 1, subfamily A, polypeptide 2 | 390 | 0.235 | 0.3244 | Yes |
| 7 | ETFDH | ETFDH Entrez, Source | electron-transferring-flavoprotein dehydrogenase | 603 | 0.186 | 0.3424 | Yes |
| 8 | NQO2 | NQO2 Entrez, Source | NAD(P)H dehydrogenase, quinone 2 | 658 | 0.179 | 0.3710 | Yes |
| 9 | ETFB | ETFB Entrez, Source | electron-transfer-flavoprotein, beta polypeptide | 746 | 0.168 | 0.3951 | Yes |
| 10 | ME1 | ME1 Entrez, Source | malic enzyme 1, NADP(+)-dependent, cytosolic | 800 | 0.163 | 0.4209 | Yes |
| 11 | AKR7A3 | AKR7A3 Entrez, Source | aldo-keto reductase family 7, member A3 (aflatoxin aldehyde reductase) | 845 | 0.158 | 0.4466 | Yes |
| 12 | UGDH | UGDH Entrez, Source | UDP-glucose dehydrogenase | 951 | 0.149 | 0.4659 | Yes |
| 13 | POR | POR Entrez, Source | P450 (cytochrome) oxidoreductase | 1030 | 0.142 | 0.4860 | Yes |
| 14 | NDUFS7 | NDUFS7 Entrez, Source | NADH dehydrogenase (ubiquinone) Fe-S protein 7, 20kDa (NADH-coenzyme Q reductase) | 1371 | 0.121 | 0.4825 | Yes |
| 15 | NQO1 | NQO1 Entrez, Source | NAD(P)H dehydrogenase, quinone 1 | 1532 | 0.112 | 0.4909 | Yes |
| 16 | QDPR | QDPR Entrez, Source | quinoid dihydropteridine reductase | 1560 | 0.110 | 0.5091 | Yes |
| 17 | MAOB | MAOB Entrez, Source | monoamine oxidase B | 1779 | 0.102 | 0.5113 | Yes |
| 18 | NDUFS1 | NDUFS1 Entrez, Source | NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) | 1817 | 0.101 | 0.5269 | Yes |
| 19 | PHYH | PHYH Entrez, Source | phytanoyl-CoA 2-hydroxylase | 2036 | 0.093 | 0.5275 | Yes |
| 20 | ADH6 | ADH6 Entrez, Source | alcohol dehydrogenase 6 (class V) | 2053 | 0.092 | 0.5432 | Yes |
| 21 | DHRS3 | DHRS3 Entrez, Source | dehydrogenase/reductase (SDR family) member 3 | 2353 | 0.084 | 0.5360 | Yes |
| 22 | SDHD | SDHD Entrez, Source | succinate dehydrogenase complex, subunit D, integral membrane protein | 2529 | 0.079 | 0.5372 | Yes |
| 23 | SRD5A1 | SRD5A1 Entrez, Source | steroid-5-alpha-reductase, alpha polypeptide 1 (3-oxo-5 alpha-steroid delta 4-dehydrogenase alpha 1) | 2561 | 0.078 | 0.5491 | Yes |
| 24 | IDH3B | IDH3B Entrez, Source | isocitrate dehydrogenase 3 (NAD+) beta | 2648 | 0.075 | 0.5564 | Yes |
| 25 | HMOX2 | HMOX2 Entrez, Source | heme oxygenase (decycling) 2 | 2666 | 0.074 | 0.5687 | Yes |
| 26 | MTRR | MTRR Entrez, Source | 5-methyltetrahydrofolate-homocysteine methyltransferase reductase | 2865 | 0.070 | 0.5666 | Yes |
| 27 | DLD | DLD Entrez, Source | dihydrolipoamide dehydrogenase | 3142 | 0.063 | 0.5574 | Yes |
| 28 | NDUFS2 | NDUFS2 Entrez, Source | NADH dehydrogenase (ubiquinone) Fe-S protein 2, 49kDa (NADH-coenzyme Q reductase) | 3172 | 0.063 | 0.5668 | Yes |
| 29 | NDUFA9 | NDUFA9 Entrez, Source | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 9, 39kDa | 3253 | 0.061 | 0.5719 | Yes |
| 30 | GSR | GSR Entrez, Source | glutathione reductase | 3579 | 0.054 | 0.5573 | No |
| 31 | NCF1 | NCF1 Entrez, Source | neutrophil cytosolic factor 1, (chronic granulomatous disease, autosomal 1) | 3743 | 0.051 | 0.5544 | No |
| 32 | PHGDH | PHGDH Entrez, Source | phosphoglycerate dehydrogenase | 4199 | 0.043 | 0.5280 | No |
| 33 | COX5A | COX5A Entrez, Source | cytochrome c oxidase subunit Va | 4442 | 0.039 | 0.5169 | No |
| 34 | CYB5R2 | CYB5R2 Entrez, Source | cytochrome b5 reductase 2 | 4527 | 0.037 | 0.5174 | No |
| 35 | GPX2 | GPX2 Entrez, Source | glutathione peroxidase 2 (gastrointestinal) | 4722 | 0.034 | 0.5090 | No |
| 36 | NDUFS4 | NDUFS4 Entrez, Source | NADH dehydrogenase (ubiquinone) Fe-S protein 4, 18kDa (NADH-coenzyme Q reductase) | 4753 | 0.033 | 0.5129 | No |
| 37 | TXNRD2 | TXNRD2 Entrez, Source | thioredoxin reductase 2 | 5055 | 0.028 | 0.4954 | No |
| 38 | ETFA | ETFA Entrez, Source | electron-transfer-flavoprotein, alpha polypeptide (glutaric aciduria II) | 5336 | 0.024 | 0.4787 | No |
| 39 | ALDH1A2 | ALDH1A2 Entrez, Source | aldehyde dehydrogenase 1 family, member A2 | 5520 | 0.021 | 0.4689 | No |
| 40 | ALDH2 | ALDH2 Entrez, Source | aldehyde dehydrogenase 2 family (mitochondrial) | 6175 | 0.012 | 0.4218 | No |
| 41 | TXNRD1 | TXNRD1 Entrez, Source | thioredoxin reductase 1 | 6256 | 0.010 | 0.4177 | No |
| 42 | FDXR | FDXR Entrez, Source | ferredoxin reductase | 6391 | 0.008 | 0.4091 | No |
| 43 | CYP19A1 | CYP19A1 Entrez, Source | cytochrome P450, family 19, subfamily A, polypeptide 1 | 6596 | 0.005 | 0.3947 | No |
| 44 | CYCS | CYCS Entrez, Source | cytochrome c, somatic | 6707 | 0.004 | 0.3871 | No |
| 45 | AKR1A1 | AKR1A1 Entrez, Source | aldo-keto reductase family 1, member A1 (aldehyde reductase) | 7127 | -0.002 | 0.3560 | No |
| 46 | CYB5R3 | CYB5R3 Entrez, Source | cytochrome b5 reductase 3 | 8007 | -0.015 | 0.2926 | No |
| 47 | NOX4 | NOX4 Entrez, Source | NADPH oxidase 4 | 8566 | -0.024 | 0.2550 | No |
| 48 | INDO | INDO Entrez, Source | indoleamine-pyrrole 2,3 dioxygenase | 9012 | -0.032 | 0.2275 | No |
| 49 | CYB5R4 | CYB5R4 Entrez, Source | cytochrome b5 reductase 4 | 11191 | -0.085 | 0.0792 | No |
| 50 | DEGS1 | DEGS1 Entrez, Source | degenerative spermatocyte homolog 1, lipid desaturase (Drosophila) | 11324 | -0.092 | 0.0861 | No |
| 51 | FDX1 | FDX1 Entrez, Source | ferredoxin 1 | 11489 | -0.099 | 0.0919 | No |
| 52 | CYBA | CYBA Entrez, Source | cytochrome b-245, alpha polypeptide | 12921 | -0.260 | 0.0317 | No |