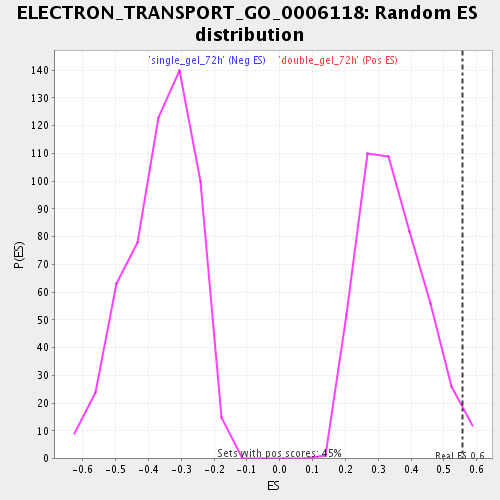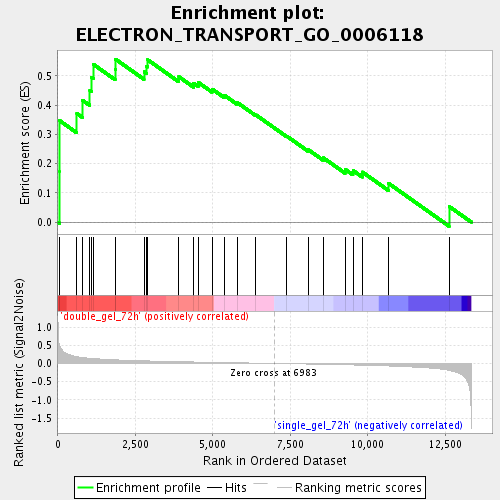
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_72h_versus_single_gel_72h.class.cls #double_gel_72h_versus_single_gel_72h_repos |
| Phenotype | class.cls#double_gel_72h_versus_single_gel_72h_repos |
| Upregulated in class | double_gel_72h |
| GeneSet | ELECTRON_TRANSPORT_GO_0006118 |
| Enrichment Score (ES) | 0.55734426 |
| Normalized Enrichment Score (NES) | 1.6153078 |
| Nominal p-value | 0.020089285 |
| FDR q-value | 0.0634647 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | ADH7 | ADH7 Entrez, Source | alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide | 53 | 0.495 | 0.1736 | Yes |
| 2 | CDO1 | CDO1 Entrez, Source | cysteine dioxygenase, type I | 58 | 0.484 | 0.3469 | Yes |
| 3 | ALDH5A1 | ALDH5A1 Entrez, Source | aldehyde dehydrogenase 5 family, member A1 (succinate-semialdehyde dehydrogenase) | 612 | 0.184 | 0.3715 | Yes |
| 4 | DHRS4 | DHRS4 Entrez, Source | dehydrogenase/reductase (SDR family) member 4 | 788 | 0.164 | 0.4172 | Yes |
| 5 | POR | POR Entrez, Source | P450 (cytochrome) oxidoreductase | 1030 | 0.142 | 0.4501 | Yes |
| 6 | CYP1B1 | CYP1B1 Entrez, Source | cytochrome P450, family 1, subfamily B, polypeptide 1 | 1078 | 0.138 | 0.4963 | Yes |
| 7 | PDIA5 | PDIA5 Entrez, Source | protein disulfide isomerase family A, member 5 | 1141 | 0.134 | 0.5398 | Yes |
| 8 | SURF1 | SURF1 Entrez, Source | surfeit 1 | 1848 | 0.100 | 0.5225 | Yes |
| 9 | AKR1D1 | AKR1D1 Entrez, Source | aldo-keto reductase family 1, member D1 (delta 4-3-ketosteroid-5-beta-reductase) | 1859 | 0.099 | 0.5573 | Yes |
| 10 | SDHA | SDHA Entrez, Source | succinate dehydrogenase complex, subunit A, flavoprotein (Fp) | 2780 | 0.072 | 0.5140 | No |
| 11 | MTRR | MTRR Entrez, Source | 5-methyltetrahydrofolate-homocysteine methyltransferase reductase | 2865 | 0.070 | 0.5328 | No |
| 12 | AOC3 | AOC3 Entrez, Source | amine oxidase, copper containing 3 (vascular adhesion protein 1) | 2897 | 0.069 | 0.5552 | No |
| 13 | UQCRC1 | UQCRC1 Entrez, Source | ubiquinol-cytochrome c reductase core protein I | 3903 | 0.048 | 0.4971 | No |
| 14 | BLVRA | BLVRA Entrez, Source | biliverdin reductase A | 4380 | 0.040 | 0.4757 | No |
| 15 | CYB5R2 | CYB5R2 Entrez, Source | cytochrome b5 reductase 2 | 4527 | 0.037 | 0.4781 | No |
| 16 | LOXL1 | LOXL1 Entrez, Source | lysyl oxidase-like 1 | 4974 | 0.030 | 0.4552 | No |
| 17 | GPX4 | GPX4 Entrez, Source | glutathione peroxidase 4 (phospholipid hydroperoxidase) | 5373 | 0.024 | 0.4338 | No |
| 18 | SDHC | SDHC Entrez, Source | succinate dehydrogenase complex, subunit C, integral membrane protein, 15kDa | 5789 | 0.018 | 0.4089 | No |
| 19 | FDXR | FDXR Entrez, Source | ferredoxin reductase | 6391 | 0.008 | 0.3666 | No |
| 20 | TXNL1 | TXNL1 Entrez, Source | thioredoxin-like 1 | 7392 | -0.006 | 0.2937 | No |
| 21 | RETSAT | RETSAT Entrez, Source | retinol saturase (all-trans-retinol 13,14-reductase) | 8072 | -0.016 | 0.2483 | No |
| 22 | NOX4 | NOX4 Entrez, Source | NADPH oxidase 4 | 8566 | -0.024 | 0.2200 | No |
| 23 | SPR | SPR Entrez, Source | sepiapterin reductase (7,8-dihydrobiopterin:NADP+ oxidoreductase) | 9268 | -0.037 | 0.1807 | No |
| 24 | DUOX2 | DUOX2 Entrez, Source | dual oxidase 2 | 9524 | -0.042 | 0.1767 | No |
| 25 | ALDH3A1 | ALDH3A1 Entrez, Source | aldehyde dehydrogenase 3 family, memberA1 | 9824 | -0.048 | 0.1716 | No |
| 26 | ALDH9A1 | ALDH9A1 Entrez, Source | aldehyde dehydrogenase 9 family, member A1 | 10674 | -0.069 | 0.1325 | No |
| 27 | PLOD1 | PLOD1 Entrez, Source | procollagen-lysine 1, 2-oxoglutarate 5-dioxygenase 1 | 12626 | -0.189 | 0.0538 | No |