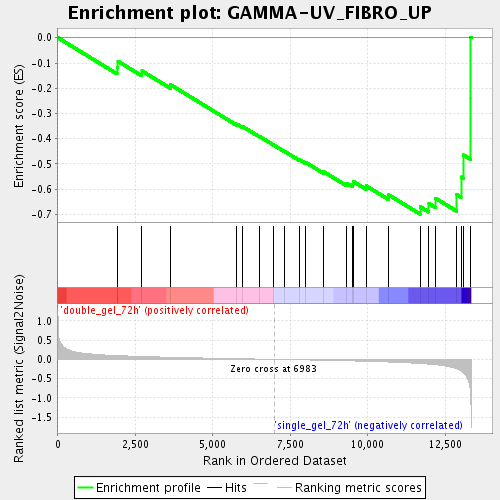
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_72h_versus_single_gel_72h.class.cls #double_gel_72h_versus_single_gel_72h_repos |
| Phenotype | class.cls#double_gel_72h_versus_single_gel_72h_repos |
| Upregulated in class | single_gel_72h |
| GeneSet | GAMMA-UV_FIBRO_UP |
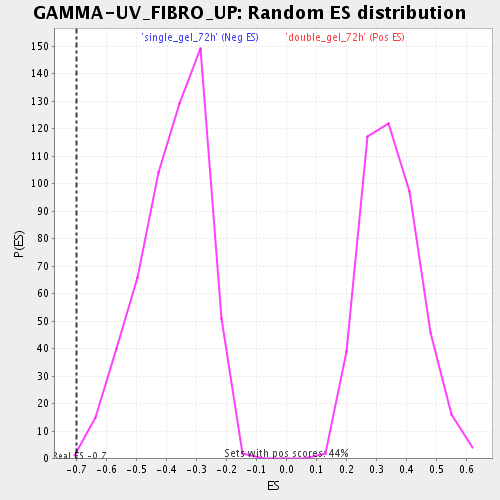
| Enrichment Score (ES) | -0.6986178 |
| Normalized Enrichment Score (NES) | -1.851004 |
| Nominal p-value | 0.0017953322 |
| FDR q-value | 0.027866052 |
| FWER p-Value | 0.806 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | CRHBP | CRHBP Entrez, Source | corticotropin releasing hormone binding protein | 1910 | 0.097 | -0.1172 | No |
| 2 | CCNH | CCNH Entrez, Source | cyclin H | 1938 | 0.096 | -0.0932 | No |
| 3 | APBA3 | APBA3 Entrez, Source | amyloid beta (A4) precursor protein-binding, family A, member 3 (X11-like 2) | 2712 | 0.073 | -0.1314 | No |
| 4 | COPB2 | COPB2 Entrez, Source | coatomer protein complex, subunit beta 2 (beta prime) | 3627 | 0.053 | -0.1856 | No |
| 5 | COG6 | COG6 Entrez, Source | component of oligomeric golgi complex 6 | 5770 | 0.018 | -0.3416 | No |
| 6 | TCF4 | TCF4 Entrez, Source | transcription factor 4 | 5953 | 0.015 | -0.3513 | No |
| 7 | RAB6A | RAB6A Entrez, Source | RAB6A, member RAS oncogene family | 6512 | 0.006 | -0.3915 | No |
| 8 | DLG1 | DLG1 Entrez, Source | discs, large homolog 1 (Drosophila) | 6958 | 0.000 | -0.4248 | No |
| 9 | ITPKB | ITPKB Entrez, Source | inositol 1,4,5-trisphosphate 3-kinase B | 7306 | -0.005 | -0.4496 | No |
| 10 | CCNC | CCNC Entrez, Source | cyclin C | 7782 | -0.012 | -0.4821 | No |
| 11 | NRP2 | NRP2 Entrez, Source | neuropilin 2 | 8004 | -0.015 | -0.4946 | No |
| 12 | CREBBP | CREBBP Entrez, Source | CREB binding protein (Rubinstein-Taybi syndrome) | 8561 | -0.024 | -0.5298 | No |
| 13 | DNAJA1 | DNAJA1 Entrez, Source | DnaJ (Hsp40) homolog, subfamily A, member 1 | 9325 | -0.038 | -0.5767 | No |
| 14 | MORF4L1 | MORF4L1 Entrez, Source | mortality factor 4 like 1 | 9516 | -0.042 | -0.5796 | No |
| 15 | CSNK1A1 | CSNK1A1 Entrez, Source | casein kinase 1, alpha 1 | 9528 | -0.042 | -0.5691 | No |
| 16 | GADD45A | GADD45A Entrez, Source | growth arrest and DNA-damage-inducible, alpha | 9946 | -0.051 | -0.5866 | No |
| 17 | ALDH9A1 | ALDH9A1 Entrez, Source | aldehyde dehydrogenase 9 family, member A1 | 10674 | -0.069 | -0.6225 | No |
| 18 | CDCA7 | CDCA7 Entrez, Source | cell division cycle associated 7 | 11689 | -0.109 | -0.6692 | Yes |
| 19 | HEXB | HEXB Entrez, Source | hexosaminidase B (beta polypeptide) | 11974 | -0.123 | -0.6572 | Yes |
| 20 | PTGER4 | PTGER4 Entrez, Source | prostaglandin E receptor 4 (subtype EP4) | 12179 | -0.137 | -0.6353 | Yes |
| 21 | ACTN4 | ACTN4 Entrez, Source | actinin, alpha 4 | 12871 | -0.243 | -0.6215 | Yes |
| 22 | TPM1 | TPM1 Entrez, Source | tropomyosin 1 (alpha) | 13011 | -0.297 | -0.5515 | Yes |
| 23 | CCND1 | CCND1 Entrez, Source | cyclin D1 | 13085 | -0.343 | -0.4643 | Yes |
| 24 | CCNA2 | CCNA2 Entrez, Source | cyclin A2 | 13314 | -0.890 | -0.2406 | Yes |
| 25 | LOX | LOX Entrez, Source | lysyl oxidase | 13315 | -0.896 | 0.0020 | Yes |