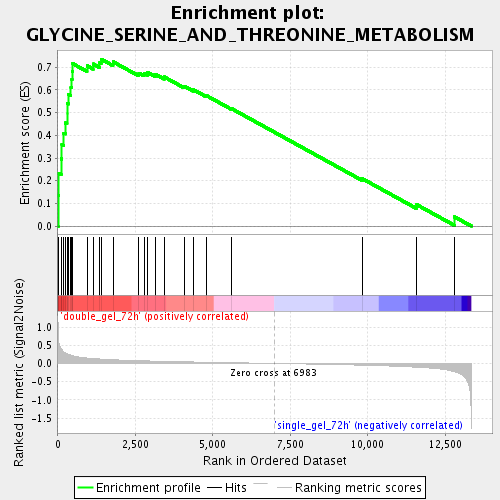
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_72h_versus_single_gel_72h.class.cls #double_gel_72h_versus_single_gel_72h_repos |
| Phenotype | class.cls#double_gel_72h_versus_single_gel_72h_repos |
| Upregulated in class | double_gel_72h |
| GeneSet | GLYCINE_SERINE_AND_THREONINE_METABOLISM |
| Enrichment Score (ES) | 0.7356597 |
| Normalized Enrichment Score (NES) | 2.156678 |
| Nominal p-value | 0.0 |
| FDR q-value | 2.0971952E-4 |
| FWER p-Value | 0.0070 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | PEMT | PEMT Entrez, Source | phosphatidylethanolamine N-methyltransferase | 8 | 0.798 | 0.1339 | Yes |
| 2 | ABP1 | ABP1 Entrez, Source | amiloride binding protein 1 (amine oxidase (copper-containing)) | 24 | 0.594 | 0.2329 | Yes |
| 3 | BHMT | BHMT Entrez, Source | betaine-homocysteine methyltransferase | 106 | 0.415 | 0.2967 | Yes |
| 4 | AGXT | AGXT Entrez, Source | alanine-glyoxylate aminotransferase (oxalosis I; hyperoxaluria I; glycolicaciduria; serine-pyruvate aminotransferase) | 128 | 0.384 | 0.3598 | Yes |
| 5 | CHDH | CHDH Entrez, Source | choline dehydrogenase | 186 | 0.321 | 0.4096 | Yes |
| 6 | SARDH | SARDH Entrez, Source | sarcosine dehydrogenase | 237 | 0.291 | 0.4549 | Yes |
| 7 | CBS | CBS Entrez, Source | cystathionine-beta-synthase | 294 | 0.267 | 0.4957 | Yes |
| 8 | GAMT | GAMT Entrez, Source | guanidinoacetate N-methyltransferase | 295 | 0.267 | 0.5408 | Yes |
| 9 | DMGDH | DMGDH Entrez, Source | dimethylglycine dehydrogenase | 334 | 0.255 | 0.5809 | Yes |
| 10 | ALAS1 | ALAS1 Entrez, Source | aminolevulinate, delta-, synthase 1 | 419 | 0.224 | 0.6125 | Yes |
| 11 | CTH | CTH Entrez, Source | cystathionase (cystathionine gamma-lyase) | 447 | 0.216 | 0.6469 | Yes |
| 12 | PSPH | PSPH Entrez, Source | phosphoserine phosphatase | 459 | 0.213 | 0.6820 | Yes |
| 13 | AGXT2 | AGXT2 Entrez, Source | alanine-glyoxylate aminotransferase 2 | 475 | 0.209 | 0.7161 | Yes |
| 14 | SHMT1 | SHMT1 Entrez, Source | serine hydroxymethyltransferase 1 (soluble) | 939 | 0.150 | 0.7066 | Yes |
| 15 | GCAT | GCAT Entrez, Source | glycine C-acetyltransferase (2-amino-3-ketobutyrate coenzyme A ligase) | 1135 | 0.135 | 0.7147 | Yes |
| 16 | SHMT2 | SHMT2 Entrez, Source | serine hydroxymethyltransferase 2 (mitochondrial) | 1347 | 0.122 | 0.7193 | Yes |
| 17 | AMT | AMT Entrez, Source | aminomethyltransferase | 1397 | 0.119 | 0.7357 | Yes |
| 18 | MAOB | MAOB Entrez, Source | monoamine oxidase B | 1779 | 0.102 | 0.7242 | No |
| 19 | CHKA | CHKA Entrez, Source | choline kinase alpha | 2612 | 0.076 | 0.6745 | No |
| 20 | PLCB2 | PLCB2 Entrez, Source | phospholipase C, beta 2 | 2782 | 0.072 | 0.6739 | No |
| 21 | AOC3 | AOC3 Entrez, Source | amine oxidase, copper containing 3 (vascular adhesion protein 1) | 2897 | 0.069 | 0.6770 | No |
| 22 | DLD | DLD Entrez, Source | dihydrolipoamide dehydrogenase | 3142 | 0.063 | 0.6694 | No |
| 23 | MAOA | MAOA Entrez, Source | monoamine oxidase A | 3427 | 0.057 | 0.6577 | No |
| 24 | TARS | TARS Entrez, Source | threonyl-tRNA synthetase | 4081 | 0.045 | 0.6162 | No |
| 25 | GARS | GARS Entrez, Source | glycyl-tRNA synthetase | 4370 | 0.040 | 0.6013 | No |
| 26 | CHKB | CHKB Entrez, Source | choline kinase beta | 4787 | 0.033 | 0.5756 | No |
| 27 | ALAS2 | ALAS2 Entrez, Source | aminolevulinate, delta-, synthase 2 (sideroblastic/hypochromic anemia) | 5606 | 0.020 | 0.5175 | No |
| 28 | PLCG2 | PLCG2 Entrez, Source | phospholipase C, gamma 2 (phosphatidylinositol-specific) | 9827 | -0.048 | 0.2087 | No |
| 29 | GATM | GATM Entrez, Source | glycine amidinotransferase (L-arginine:glycine amidinotransferase) | 11580 | -0.103 | 0.0945 | No |
| 30 | PLCG1 | PLCG1 Entrez, Source | phospholipase C, gamma 1 | 12798 | -0.224 | 0.0409 | No |

