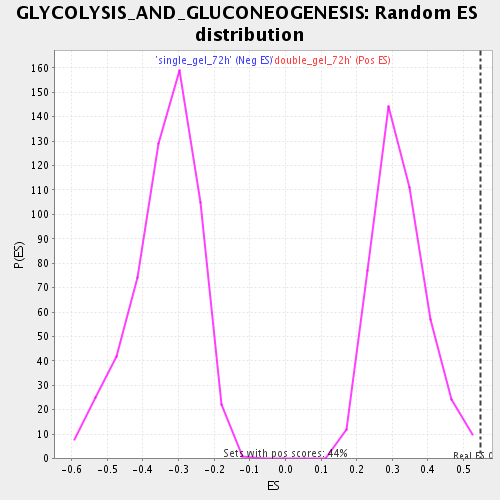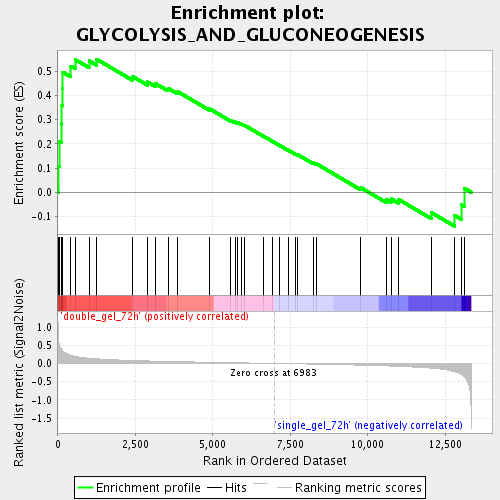
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_72h_versus_single_gel_72h.class.cls #double_gel_72h_versus_single_gel_72h_repos |
| Phenotype | class.cls#double_gel_72h_versus_single_gel_72h_repos |
| Upregulated in class | double_gel_72h |
| GeneSet | GLYCOLYSIS_AND_GLUCONEOGENESIS |
| Enrichment Score (ES) | 0.54913473 |
| Normalized Enrichment Score (NES) | 1.7081558 |
| Nominal p-value | 0.0022988506 |
| FDR q-value | 0.03474909 |
| FWER p-Value | 0.997 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | PKLR | PKLR Entrez, Source | pyruvate kinase, liver and RBC | 31 | 0.561 | 0.1079 | Yes |
| 2 | FBP1 | FBP1 Entrez, Source | fructose-1,6-bisphosphatase 1 | 43 | 0.515 | 0.2084 | Yes |
| 3 | GOT1 | GOT1 Entrez, Source | glutamic-oxaloacetic transaminase 1, soluble (aspartate aminotransferase 1) | 110 | 0.403 | 0.2826 | Yes |
| 4 | G6PC | G6PC Entrez, Source | glucose-6-phosphatase, catalytic subunit | 129 | 0.383 | 0.3567 | Yes |
| 5 | GCK | GCK Entrez, Source | glucokinase (hexokinase 4, maturity onset diabetes of the young 2) | 140 | 0.368 | 0.4283 | Yes |
| 6 | PC | PC Entrez, Source | pyruvate carboxylase | 155 | 0.347 | 0.4955 | Yes |
| 7 | PDHA2 | PDHA2 Entrez, Source | pyruvate dehydrogenase (lipoamide) alpha 2 | 414 | 0.227 | 0.5207 | Yes |
| 8 | PDHB | PDHB Entrez, Source | pyruvate dehydrogenase (lipoamide) beta | 563 | 0.190 | 0.5471 | Yes |
| 9 | ALDOB | ALDOB Entrez, Source | aldolase B, fructose-bisphosphate | 1007 | 0.144 | 0.5421 | Yes |
| 10 | MDH1 | MDH1 Entrez, Source | malate dehydrogenase 1, NAD (soluble) | 1246 | 0.127 | 0.5491 | Yes |
| 11 | MDH2 | MDH2 Entrez, Source | malate dehydrogenase 2, NAD (mitochondrial) | 2409 | 0.082 | 0.4780 | No |
| 12 | ENO2 | ENO2 Entrez, Source | enolase 2 (gamma, neuronal) | 2893 | 0.069 | 0.4553 | No |
| 13 | DLD | DLD Entrez, Source | dihydrolipoamide dehydrogenase | 3142 | 0.063 | 0.4491 | No |
| 14 | HK2 | HK2 Entrez, Source | hexokinase 2 | 3553 | 0.055 | 0.4291 | No |
| 15 | ENO3 | ENO3 Entrez, Source | enolase 3 (beta, muscle) | 3849 | 0.049 | 0.4165 | No |
| 16 | FBP2 | FBP2 Entrez, Source | fructose-1,6-bisphosphatase 2 | 4892 | 0.031 | 0.3443 | No |
| 17 | GPI | GPI Entrez, Source | glucose phosphate isomerase | 5582 | 0.021 | 0.2966 | No |
| 18 | ALDOC | ALDOC Entrez, Source | aldolase C, fructose-bisphosphate | 5718 | 0.018 | 0.2901 | No |
| 19 | PDHA1 | PDHA1 Entrez, Source | pyruvate dehydrogenase (lipoamide) alpha 1 | 5797 | 0.017 | 0.2877 | No |
| 20 | GOT2 | GOT2 Entrez, Source | glutamic-oxaloacetic transaminase 2, mitochondrial (aspartate aminotransferase 2) | 5916 | 0.015 | 0.2818 | No |
| 21 | PCK1 | PCK1 Entrez, Source | phosphoenolpyruvate carboxykinase 1 (soluble) | 6026 | 0.014 | 0.2763 | No |
| 22 | LDHC | LDHC Entrez, Source | lactate dehydrogenase C | 6640 | 0.005 | 0.2312 | No |
| 23 | GAPDHS | GAPDHS Entrez, Source | glyceraldehyde-3-phosphate dehydrogenase, spermatogenic | 6929 | 0.001 | 0.2097 | No |
| 24 | PGK1 | PGK1 Entrez, Source | phosphoglycerate kinase 1 | 7152 | -0.002 | 0.1935 | No |
| 25 | PGAM2 | PGAM2 Entrez, Source | phosphoglycerate mutase 2 (muscle) | 7429 | -0.006 | 0.1740 | No |
| 26 | LDHA | LDHA Entrez, Source | lactate dehydrogenase A | 7676 | -0.010 | 0.1575 | No |
| 27 | GAPDH | GAPDH Entrez, Source | glyceraldehyde-3-phosphate dehydrogenase | 7743 | -0.011 | 0.1547 | No |
| 28 | TPI1 | TPI1 Entrez, Source | triosephosphate isomerase 1 | 8236 | -0.019 | 0.1214 | No |
| 29 | LDHAL6B | LDHAL6B Entrez, Source | lactate dehydrogenase A-like 6B | 8335 | -0.020 | 0.1181 | No |
| 30 | ALDOA | ALDOA Entrez, Source | aldolase A, fructose-bisphosphate | 9779 | -0.047 | 0.0190 | No |
| 31 | PGAM1 | PGAM1 Entrez, Source | phosphoglycerate mutase 1 (brain) | 10612 | -0.067 | -0.0303 | No |
| 32 | PFKL | PFKL Entrez, Source | phosphofructokinase, liver | 10756 | -0.071 | -0.0270 | No |
| 33 | DLAT | DLAT Entrez, Source | dihydrolipoamide S-acetyltransferase (E2 component of pyruvate dehydrogenase complex) | 10995 | -0.078 | -0.0295 | No |
| 34 | HK1 | HK1 Entrez, Source | hexokinase 1 | 12060 | -0.129 | -0.0842 | No |
| 35 | PFKM | PFKM Entrez, Source | phosphofructokinase, muscle | 12804 | -0.226 | -0.0956 | No |
| 36 | LDHB | LDHB Entrez, Source | lactate dehydrogenase B | 13034 | -0.309 | -0.0519 | No |
| 37 | PFKP | PFKP Entrez, Source | phosphofructokinase, platelet | 13129 | -0.381 | 0.0160 | No |