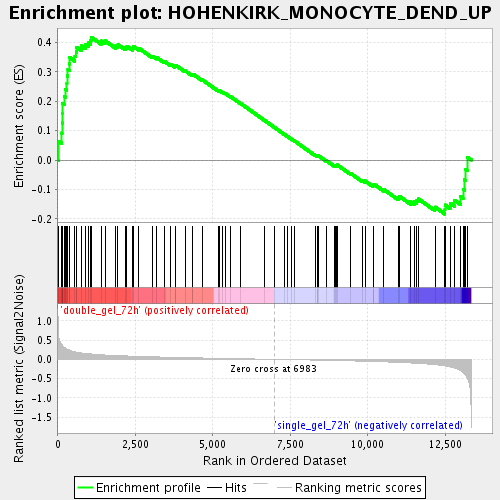
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_72h_versus_single_gel_72h.class.cls #double_gel_72h_versus_single_gel_72h_repos |
| Phenotype | class.cls#double_gel_72h_versus_single_gel_72h_repos |
| Upregulated in class | double_gel_72h |
| GeneSet | HOHENKIRK_MONOCYTE_DEND_UP |
| Enrichment Score (ES) | 0.41748965 |
| Normalized Enrichment Score (NES) | 1.5182601 |
| Nominal p-value | 0.005063291 |
| FDR q-value | 0.1112523 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | FCGR2B | FCGR2B Entrez, Source | Fc fragment of IgG, low affinity IIb, receptor (CD32) | 16 | 0.713 | 0.0648 | Yes |
| 2 | APOC1 | APOC1 Entrez, Source | apolipoprotein C-I | 127 | 0.384 | 0.0920 | Yes |
| 3 | ACAA2 | ACAA2 Entrez, Source | acetyl-Coenzyme A acyltransferase 2 (mitochondrial 3-oxoacyl-Coenzyme A thiolase) | 135 | 0.374 | 0.1262 | Yes |
| 4 | ENPP2 | ENPP2 Entrez, Source | ectonucleotide pyrophosphatase/phosphodiesterase 2 (autotaxin) | 143 | 0.363 | 0.1593 | Yes |
| 5 | SULT1C1 | SULT1C1 Entrez, Source | sulfotransferase family, cytosolic, 1C, member 1 | 148 | 0.356 | 0.1920 | Yes |
| 6 | CEBPA | CEBPA Entrez, Source | CCAAT/enhancer binding protein (C/EBP), alpha | 203 | 0.308 | 0.2164 | Yes |
| 7 | RARRES1 | RARRES1 Entrez, Source | retinoic acid receptor responder (tazarotene induced) 1 | 239 | 0.290 | 0.2405 | Yes |
| 8 | CAT | CAT Entrez, Source | catalase | 290 | 0.269 | 0.2617 | Yes |
| 9 | CBS | CBS Entrez, Source | cystathionine-beta-synthase | 294 | 0.267 | 0.2862 | Yes |
| 10 | SLC11A2 | SLC11A2 Entrez, Source | solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2 | 322 | 0.259 | 0.3082 | Yes |
| 11 | HSD11B1 | HSD11B1 Entrez, Source | hydroxysteroid (11-beta) dehydrogenase 1 | 370 | 0.242 | 0.3270 | Yes |
| 12 | C1QB | C1QB Entrez, Source | complement component 1, q subcomponent, B chain | 383 | 0.238 | 0.3482 | Yes |
| 13 | TNFAIP6 | TNFAIP6 Entrez, Source | tumor necrosis factor, alpha-induced protein 6 | 548 | 0.192 | 0.3536 | Yes |
| 14 | NR1H3 | NR1H3 Entrez, Source | nuclear receptor subfamily 1, group H, member 3 | 583 | 0.188 | 0.3684 | Yes |
| 15 | TFRC | TFRC Entrez, Source | transferrin receptor (p90, CD71) | 608 | 0.185 | 0.3838 | Yes |
| 16 | MGLL | MGLL Entrez, Source | monoglyceride lipase | 754 | 0.167 | 0.3883 | Yes |
| 17 | OAT | OAT Entrez, Source | ornithine aminotransferase (gyrate atrophy) | 893 | 0.154 | 0.3921 | Yes |
| 18 | TGFA | TGFA Entrez, Source | transforming growth factor, alpha | 986 | 0.146 | 0.3987 | Yes |
| 19 | CD86 | CD86 Entrez, Source | CD86 molecule | 1061 | 0.139 | 0.4060 | Yes |
| 20 | SLA | SLA Entrez, Source | Src-like-adaptor | 1080 | 0.138 | 0.4175 | Yes |
| 21 | ST6GAL1 | ST6GAL1 Entrez, Source | ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 | 1393 | 0.119 | 0.4050 | No |
| 22 | CD74 | CD74 Entrez, Source | CD74 molecule, major histocompatibility complex, class II invariant chain | 1524 | 0.112 | 0.4056 | No |
| 23 | PCBD1 | PCBD1 Entrez, Source | pterin-4 alpha-carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha (TCF1) | 1870 | 0.099 | 0.3887 | No |
| 24 | CCNH | CCNH Entrez, Source | cyclin H | 1938 | 0.096 | 0.3925 | No |
| 25 | CYB5A | CYB5A Entrez, Source | cytochrome b5 type A (microsomal) | 2179 | 0.089 | 0.3826 | No |
| 26 | DUSP5 | DUSP5 Entrez, Source | dual specificity phosphatase 5 | 2225 | 0.087 | 0.3873 | No |
| 27 | PTPRO | PTPRO Entrez, Source | protein tyrosine phosphatase, receptor type, O | 2417 | 0.082 | 0.3805 | No |
| 28 | CCL22 | CCL22 Entrez, Source | chemokine (C-C motif) ligand 22 | 2442 | 0.081 | 0.3862 | No |
| 29 | DPP4 | DPP4 Entrez, Source | dipeptidyl-peptidase 4 (CD26, adenosine deaminase complexing protein 2) | 2611 | 0.076 | 0.3806 | No |
| 30 | ACOT7 | ACOT7 Entrez, Source | acyl-CoA thioesterase 7 | 3055 | 0.065 | 0.3532 | No |
| 31 | MMP12 | MMP12 Entrez, Source | matrix metallopeptidase 12 (macrophage elastase) | 3186 | 0.063 | 0.3492 | No |
| 32 | MAOA | MAOA Entrez, Source | monoamine oxidase A | 3427 | 0.057 | 0.3364 | No |
| 33 | ACOT2 | ACOT2 Entrez, Source | acyl-CoA thioesterase 2 | 3642 | 0.053 | 0.3252 | No |
| 34 | DNASE1L3 | DNASE1L3 Entrez, Source | deoxyribonuclease I-like 3 | 3779 | 0.050 | 0.3196 | No |
| 35 | CTSC | CTSC Entrez, Source | cathepsin C | 3796 | 0.050 | 0.3230 | No |
| 36 | CCL17 | CCL17 Entrez, Source | chemokine (C-C motif) ligand 17 | 4105 | 0.045 | 0.3039 | No |
| 37 | SPP1 | SPP1 Entrez, Source | secreted phosphoprotein 1 (osteopontin, bone sialoprotein I, early T-lymphocyte activation 1) | 4327 | 0.041 | 0.2910 | No |
| 38 | PDLIM4 | PDLIM4 Entrez, Source | PDZ and LIM domain 4 | 4357 | 0.040 | 0.2925 | No |
| 39 | GAS6 | GAS6 Entrez, Source | growth arrest-specific 6 | 4656 | 0.035 | 0.2733 | No |
| 40 | PEBP1 | PEBP1 Entrez, Source | phosphatidylethanolamine binding protein 1 | 5190 | 0.026 | 0.2356 | No |
| 41 | TOMM40 | TOMM40 Entrez, Source | translocase of outer mitochondrial membrane 40 homolog (yeast) | 5228 | 0.026 | 0.2351 | No |
| 42 | CDH1 | CDH1 Entrez, Source | cadherin 1, type 1, E-cadherin (epithelial) | 5315 | 0.024 | 0.2309 | No |
| 43 | ALOX15 | ALOX15 Entrez, Source | arachidonate 15-lipoxygenase | 5400 | 0.023 | 0.2267 | No |
| 44 | A2M | A2M Entrez, Source | alpha-2-macroglobulin | 5584 | 0.021 | 0.2148 | No |
| 45 | CSF1 | CSF1 Entrez, Source | colony stimulating factor 1 (macrophage) | 5899 | 0.016 | 0.1926 | No |
| 46 | DUSP1 | DUSP1 Entrez, Source | dual specificity phosphatase 1 | 6673 | 0.004 | 0.1347 | No |
| 47 | INHBA | INHBA Entrez, Source | inhibin, beta A (activin A, activin AB alpha polypeptide) | 6984 | -0.000 | 0.1113 | No |
| 48 | ITPKB | ITPKB Entrez, Source | inositol 1,4,5-trisphosphate 3-kinase B | 7306 | -0.005 | 0.0876 | No |
| 49 | ATP1B1 | ATP1B1 Entrez, Source | ATPase, Na+/K+ transporting, beta 1 polypeptide | 7404 | -0.006 | 0.0808 | No |
| 50 | CRABP2 | CRABP2 Entrez, Source | cellular retinoic acid binding protein 2 | 7542 | -0.008 | 0.0712 | No |
| 51 | NR4A3 | NR4A3 Entrez, Source | nuclear receptor subfamily 4, group A, member 3 | 7647 | -0.010 | 0.0643 | No |
| 52 | ITGB5 | ITGB5 Entrez, Source | integrin, beta 5 | 8308 | -0.020 | 0.0164 | No |
| 53 | RNASE6 | RNASE6 Entrez, Source | ribonuclease, RNase A family, k6 | 8373 | -0.021 | 0.0135 | No |
| 54 | CCNA1 | CCNA1 Entrez, Source | cyclin A1 | 8384 | -0.021 | 0.0147 | No |
| 55 | SNRPN | SNRPN Entrez, Source | small nuclear ribonucleoprotein polypeptide N | 8412 | -0.022 | 0.0147 | No |
| 56 | SPINT2 | SPINT2 Entrez, Source | serine peptidase inhibitor, Kunitz type, 2 | 8659 | -0.026 | -0.0015 | No |
| 57 | F13A1 | F13A1 Entrez, Source | coagulation factor XIII, A1 polypeptide | 8934 | -0.031 | -0.0193 | No |
| 58 | HMGN3 | HMGN3 Entrez, Source | high mobility group nucleosomal binding domain 3 | 8947 | -0.031 | -0.0173 | No |
| 59 | KCNAB2 | KCNAB2 Entrez, Source | potassium voltage-gated channel, shaker-related subfamily, beta member 2 | 9000 | -0.032 | -0.0183 | No |
| 60 | INDO | INDO Entrez, Source | indoleamine-pyrrole 2,3 dioxygenase | 9012 | -0.032 | -0.0161 | No |
| 61 | FABP4 | FABP4 Entrez, Source | fatty acid binding protein 4, adipocyte | 9450 | -0.041 | -0.0453 | No |
| 62 | FABP5 | FABP5 Entrez, Source | fatty acid binding protein 5 (psoriasis-associated) | 9829 | -0.048 | -0.0693 | No |
| 63 | ARF4 | ARF4 Entrez, Source | ADP-ribosylation factor 4 | 9911 | -0.050 | -0.0708 | No |
| 64 | TGM2 | TGM2 Entrez, Source | transglutaminase 2 (C polypeptide, protein-glutamine-gamma-glutamyltransferase) | 10171 | -0.056 | -0.0851 | No |
| 65 | AES | AES Entrez, Source | amino-terminal enhancer of split | 10196 | -0.057 | -0.0817 | No |
| 66 | ECM1 | ECM1 Entrez, Source | extracellular matrix protein 1 | 10516 | -0.065 | -0.0998 | No |
| 67 | LPL | LPL Entrez, Source | lipoprotein lipase | 10988 | -0.078 | -0.1281 | No |
| 68 | GDF15 | GDF15 Entrez, Source | growth differentiation factor 15 | 11020 | -0.079 | -0.1231 | No |
| 69 | RAP1GAP | RAP1GAP Entrez, Source | RAP1 GTPase activating protein | 11386 | -0.094 | -0.1419 | No |
| 70 | SCARB1 | SCARB1 Entrez, Source | scavenger receptor class B, member 1 | 11498 | -0.099 | -0.1410 | No |
| 71 | GATM | GATM Entrez, Source | glycine amidinotransferase (L-arginine:glycine amidinotransferase) | 11580 | -0.103 | -0.1376 | No |
| 72 | BIRC3 | BIRC3 Entrez, Source | baculoviral IAP repeat-containing 3 | 11637 | -0.106 | -0.1320 | No |
| 73 | QSCN6 | QSCN6 Entrez, Source | quiescin Q6 | 12170 | -0.136 | -0.1595 | No |
| 74 | CD59 | CD59 Entrez, Source | CD59 molecule, complement regulatory protein | 12491 | -0.169 | -0.1680 | No |
| 75 | LGALS3BP | LGALS3BP Entrez, Source | lectin, galactoside-binding, soluble, 3 binding protein | 12495 | -0.170 | -0.1525 | No |
| 76 | PPIC | PPIC Entrez, Source | peptidylprolyl isomerase C (cyclophilin C) | 12659 | -0.196 | -0.1467 | No |
| 77 | PFKM | PFKM Entrez, Source | phosphofructokinase, muscle | 12804 | -0.226 | -0.1366 | No |
| 78 | FABP3 | FABP3 Entrez, Source | fatty acid binding protein 3, muscle and heart (mammary-derived growth inhibitor) | 12998 | -0.290 | -0.1243 | No |
| 79 | CCND2 | CCND2 Entrez, Source | cyclin D2 | 13080 | -0.340 | -0.0989 | No |
| 80 | PFKP | PFKP Entrez, Source | phosphofructokinase, platelet | 13129 | -0.381 | -0.0672 | No |
| 81 | ABCG1 | ABCG1 Entrez, Source | ATP-binding cassette, sub-family G (WHITE), member 1 | 13162 | -0.417 | -0.0310 | No |
| 82 | CITED2 | CITED2 Entrez, Source | Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 | 13211 | -0.481 | 0.0099 | No |

