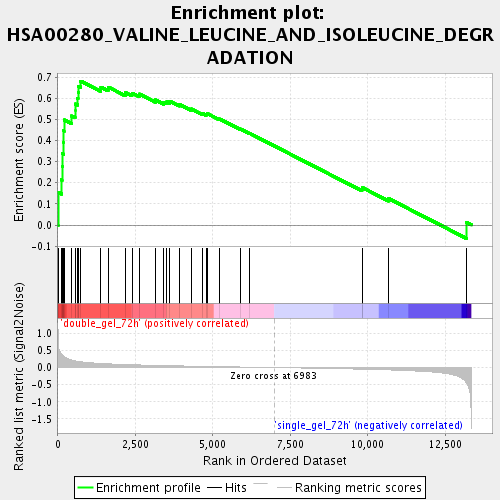
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_72h_versus_single_gel_72h.class.cls #double_gel_72h_versus_single_gel_72h_repos |
| Phenotype | class.cls#double_gel_72h_versus_single_gel_72h_repos |
| Upregulated in class | double_gel_72h |
| GeneSet | HSA00280_VALINE_LEUCINE_AND_ISOLEUCINE_DEGRADATION |
| Enrichment Score (ES) | 0.68079877 |
| Normalized Enrichment Score (NES) | 2.0978806 |
| Nominal p-value | 0.0 |
| FDR q-value | 6.170183E-4 |
| FWER p-Value | 0.023 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | EHHADH | EHHADH Entrez, Source | enoyl-Coenzyme A, hydratase/3-hydroxyacyl Coenzyme A dehydrogenase | 6 | 0.908 | 0.1544 | Yes |
| 2 | HMGCS2 | HMGCS2 Entrez, Source | 3-hydroxy-3-methylglutaryl-Coenzyme A synthase 2 (mitochondrial) | 117 | 0.396 | 0.2137 | Yes |
| 3 | ACAA2 | ACAA2 Entrez, Source | acetyl-Coenzyme A acyltransferase 2 (mitochondrial 3-oxoacyl-Coenzyme A thiolase) | 135 | 0.374 | 0.2763 | Yes |
| 4 | ACAT2 | ACAT2 Entrez, Source | acetyl-Coenzyme A acetyltransferase 2 (acetoacetyl Coenzyme A thiolase) | 144 | 0.362 | 0.3374 | Yes |
| 5 | HMGCS1 | HMGCS1 Entrez, Source | 3-hydroxy-3-methylglutaryl-Coenzyme A synthase 1 (soluble) | 178 | 0.330 | 0.3911 | Yes |
| 6 | ABAT | ABAT Entrez, Source | 4-aminobutyrate aminotransferase | 185 | 0.321 | 0.4455 | Yes |
| 7 | AOX1 | AOX1 Entrez, Source | aldehyde oxidase 1 | 201 | 0.309 | 0.4971 | Yes |
| 8 | ALDH6A1 | ALDH6A1 Entrez, Source | aldehyde dehydrogenase 6 family, member A1 | 445 | 0.217 | 0.5159 | Yes |
| 9 | MCCC1 | MCCC1 Entrez, Source | methylcrotonoyl-Coenzyme A carboxylase 1 (alpha) | 556 | 0.192 | 0.5403 | Yes |
| 10 | ACAT1 | ACAT1 Entrez, Source | acetyl-Coenzyme A acetyltransferase 1 (acetoacetyl Coenzyme A thiolase) | 560 | 0.191 | 0.5726 | Yes |
| 11 | ECHS1 | ECHS1 Entrez, Source | enoyl Coenzyme A hydratase, short chain, 1, mitochondrial | 633 | 0.182 | 0.5983 | Yes |
| 12 | BCKDHB | BCKDHB Entrez, Source | branched chain keto acid dehydrogenase E1, beta polypeptide (maple syrup urine disease) | 651 | 0.179 | 0.6275 | Yes |
| 13 | BCKDHA | BCKDHA Entrez, Source | branched chain keto acid dehydrogenase E1, alpha polypeptide | 662 | 0.178 | 0.6572 | Yes |
| 14 | HMGCL | HMGCL Entrez, Source | 3-hydroxymethyl-3-methylglutaryl-Coenzyme A lyase (hydroxymethylglutaricaciduria) | 732 | 0.169 | 0.6808 | Yes |
| 15 | HIBADH | HIBADH Entrez, Source | 3-hydroxyisobutyrate dehydrogenase | 1384 | 0.120 | 0.6523 | No |
| 16 | HADH | HADH Entrez, Source | hydroxyacyl-Coenzyme A dehydrogenase | 1622 | 0.108 | 0.6529 | No |
| 17 | HADHA | HADHA Entrez, Source | hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (trifunctional protein), alpha subunit | 2174 | 0.089 | 0.6266 | No |
| 18 | HIBCH | HIBCH Entrez, Source | 3-hydroxyisobutyryl-Coenzyme A hydrolase | 2392 | 0.083 | 0.6244 | No |
| 19 | HSD17B4 | HSD17B4 Entrez, Source | hydroxysteroid (17-beta) dehydrogenase 4 | 2625 | 0.076 | 0.6199 | No |
| 20 | DLD | DLD Entrez, Source | dihydrolipoamide dehydrogenase | 3142 | 0.063 | 0.5919 | No |
| 21 | PCCB | PCCB Entrez, Source | propionyl Coenzyme A carboxylase, beta polypeptide | 3417 | 0.058 | 0.5812 | No |
| 22 | MCCC2 | MCCC2 Entrez, Source | methylcrotonoyl-Coenzyme A carboxylase 2 (beta) | 3517 | 0.056 | 0.5832 | No |
| 23 | ALDH7A1 | ALDH7A1 Entrez, Source | aldehyde dehydrogenase 7 family, member A1 | 3614 | 0.053 | 0.5851 | No |
| 24 | DBT | DBT Entrez, Source | dihydrolipoamide branched chain transacylase E2 | 3912 | 0.048 | 0.5710 | No |
| 25 | ACADM | ACADM Entrez, Source | acyl-Coenzyme A dehydrogenase, C-4 to C-12 straight chain | 4299 | 0.041 | 0.5490 | No |
| 26 | HADHB | HADHB Entrez, Source | hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (trifunctional protein), beta subunit | 4675 | 0.035 | 0.5268 | No |
| 27 | IVD | IVD Entrez, Source | isovaleryl Coenzyme A dehydrogenase | 4783 | 0.033 | 0.5243 | No |
| 28 | ALDH3A2 | ALDH3A2 Entrez, Source | aldehyde dehydrogenase 3 family, member A2 | 4819 | 0.032 | 0.5272 | No |
| 29 | ALDH1B1 | ALDH1B1 Entrez, Source | aldehyde dehydrogenase 1 family, member B1 | 5218 | 0.026 | 0.5017 | No |
| 30 | ACADS | ACADS Entrez, Source | acyl-Coenzyme A dehydrogenase, C-2 to C-3 short chain | 5886 | 0.016 | 0.4543 | No |
| 31 | ALDH2 | ALDH2 Entrez, Source | aldehyde dehydrogenase 2 family (mitochondrial) | 6175 | 0.012 | 0.4347 | No |
| 32 | BCAT2 | BCAT2 Entrez, Source | branched chain aminotransferase 2, mitochondrial | 9815 | -0.048 | 0.1694 | No |
| 33 | ALDH3A1 | ALDH3A1 Entrez, Source | aldehyde dehydrogenase 3 family, memberA1 | 9824 | -0.048 | 0.1771 | No |
| 34 | ALDH9A1 | ALDH9A1 Entrez, Source | aldehyde dehydrogenase 9 family, member A1 | 10674 | -0.069 | 0.1251 | No |
| 35 | BCAT1 | BCAT1 Entrez, Source | branched chain aminotransferase 1, cytosolic | 13181 | -0.442 | 0.0121 | No |