Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_72h_versus_single_gel_72h.class.cls #double_gel_72h_versus_single_gel_72h_repos |
| Phenotype | class.cls#double_gel_72h_versus_single_gel_72h_repos |
| Upregulated in class | double_gel_72h |
| GeneSet | HSA00340_HISTIDINE_METABOLISM |
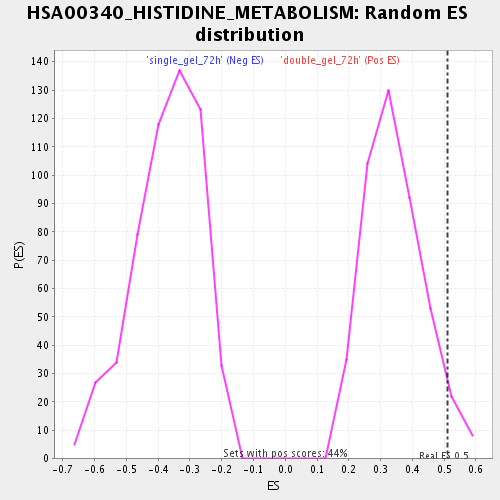
| Enrichment Score (ES) | 0.51034784 |
| Normalized Enrichment Score (NES) | 1.4914336 |
| Nominal p-value | 0.04954955 |
| FDR q-value | 0.12545732 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | FTCD | FTCD Entrez, Source | formiminotransferase cyclodeaminase | 15 | 0.731 | 0.2263 | Yes |
| 2 | ABP1 | ABP1 Entrez, Source | amiloride binding protein 1 (amine oxidase (copper-containing)) | 24 | 0.594 | 0.4102 | Yes |
| 3 | HAL | HAL Entrez, Source | histidine ammonia-lyase | 423 | 0.224 | 0.4500 | Yes |
| 4 | PRMT3 | PRMT3 Entrez, Source | protein arginine methyltransferase 3 | 498 | 0.203 | 0.5076 | Yes |
| 5 | HDC | HDC Entrez, Source | histidine decarboxylase | 1527 | 0.112 | 0.4652 | Yes |
| 6 | ASPA | ASPA Entrez, Source | aspartoacylase (Canavan disease) | 1733 | 0.104 | 0.4821 | Yes |
| 7 | MAOB | MAOB Entrez, Source | monoamine oxidase B | 1779 | 0.102 | 0.5103 | Yes |
| 8 | AOC3 | AOC3 Entrez, Source | amine oxidase, copper containing 3 (vascular adhesion protein 1) | 2897 | 0.069 | 0.4479 | No |
| 9 | MAOA | MAOA Entrez, Source | monoamine oxidase A | 3427 | 0.057 | 0.4260 | No |
| 10 | ALDH7A1 | ALDH7A1 Entrez, Source | aldehyde dehydrogenase 7 family, member A1 | 3614 | 0.053 | 0.4287 | No |
| 11 | LCMT2 | LCMT2 Entrez, Source | leucine carboxyl methyltransferase 2 | 3734 | 0.051 | 0.4356 | No |
| 12 | ALDH3A2 | ALDH3A2 Entrez, Source | aldehyde dehydrogenase 3 family, member A2 | 4819 | 0.032 | 0.3642 | No |
| 13 | ALDH1B1 | ALDH1B1 Entrez, Source | aldehyde dehydrogenase 1 family, member B1 | 5218 | 0.026 | 0.3424 | No |
| 14 | LCMT1 | LCMT1 Entrez, Source | leucine carboxyl methyltransferase 1 | 5536 | 0.021 | 0.3252 | No |
| 15 | ALDH2 | ALDH2 Entrez, Source | aldehyde dehydrogenase 2 family (mitochondrial) | 6175 | 0.012 | 0.2809 | No |
| 16 | PRPS1 | PRPS1 Entrez, Source | phosphoribosyl pyrophosphate synthetase 1 | 6821 | 0.002 | 0.2332 | No |
| 17 | DDC | DDC Entrez, Source | dopa decarboxylase (aromatic L-amino acid decarboxylase) | 8401 | -0.021 | 0.1213 | No |
| 18 | METTL6 | METTL6 Entrez, Source | methyltransferase like 6 | 8832 | -0.029 | 0.0979 | No |
| 19 | CARM1 | CARM1 Entrez, Source | coactivator-associated arginine methyltransferase 1 | 9227 | -0.036 | 0.0796 | No |
| 20 | ACY3 | ACY3 Entrez, Source | aspartoacylase (aminocyclase) 3 | 9233 | -0.036 | 0.0905 | No |
| 21 | CNDP1 | CNDP1 Entrez, Source | carnosine dipeptidase 1 (metallopeptidase M20 family) | 9246 | -0.037 | 0.1011 | No |
| 22 | ALDH3A1 | ALDH3A1 Entrez, Source | aldehyde dehydrogenase 3 family, memberA1 | 9824 | -0.048 | 0.0728 | No |
| 23 | PRMT2 | PRMT2 Entrez, Source | protein arginine methyltransferase 2 | 9939 | -0.051 | 0.0800 | No |
| 24 | ALDH9A1 | ALDH9A1 Entrez, Source | aldehyde dehydrogenase 9 family, member A1 | 10674 | -0.069 | 0.0464 | No |
| 25 | HNMT | HNMT Entrez, Source | histamine N-methyltransferase | 11747 | -0.111 | 0.0004 | No |
| 26 | PRPS2 | PRPS2 Entrez, Source | phosphoribosyl pyrophosphate synthetase 2 | 12448 | -0.164 | -0.0012 | No |
| 27 | ALDH3B1 | ALDH3B1 Entrez, Source | aldehyde dehydrogenase 3 family, member B1 | 12784 | -0.220 | 0.0419 | No |