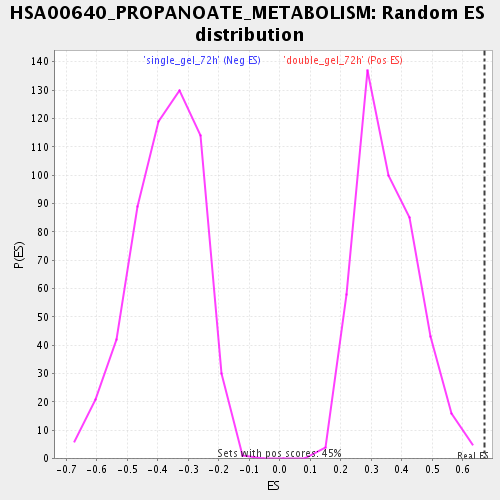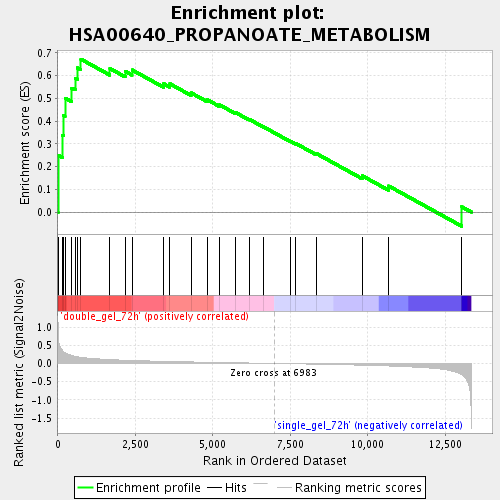
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_72h_versus_single_gel_72h.class.cls #double_gel_72h_versus_single_gel_72h_repos |
| Phenotype | class.cls#double_gel_72h_versus_single_gel_72h_repos |
| Upregulated in class | double_gel_72h |
| GeneSet | HSA00640_PROPANOATE_METABOLISM |
| Enrichment Score (ES) | 0.67272455 |
| Normalized Enrichment Score (NES) | 1.9052469 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.006388329 |
| FWER p-Value | 0.433 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | EHHADH | EHHADH Entrez, Source | enoyl-Coenzyme A, hydratase/3-hydroxyacyl Coenzyme A dehydrogenase | 6 | 0.908 | 0.2497 | Yes |
| 2 | ACAT2 | ACAT2 Entrez, Source | acetyl-Coenzyme A acetyltransferase 2 (acetoacetyl Coenzyme A thiolase) | 144 | 0.362 | 0.3390 | Yes |
| 3 | ABAT | ABAT Entrez, Source | 4-aminobutyrate aminotransferase | 185 | 0.321 | 0.4246 | Yes |
| 4 | ACACA | ACACA Entrez, Source | acetyl-Coenzyme A carboxylase alpha | 231 | 0.293 | 0.5019 | Yes |
| 5 | ALDH6A1 | ALDH6A1 Entrez, Source | aldehyde dehydrogenase 6 family, member A1 | 445 | 0.217 | 0.5458 | Yes |
| 6 | ACAT1 | ACAT1 Entrez, Source | acetyl-Coenzyme A acetyltransferase 1 (acetoacetyl Coenzyme A thiolase) | 560 | 0.191 | 0.5898 | Yes |
| 7 | ECHS1 | ECHS1 Entrez, Source | enoyl Coenzyme A hydratase, short chain, 1, mitochondrial | 633 | 0.182 | 0.6345 | Yes |
| 8 | ACACB | ACACB Entrez, Source | acetyl-Coenzyme A carboxylase beta | 742 | 0.168 | 0.6727 | Yes |
| 9 | MLYCD | MLYCD Entrez, Source | malonyl-CoA decarboxylase | 1678 | 0.106 | 0.6317 | No |
| 10 | HADHA | HADHA Entrez, Source | hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (trifunctional protein), alpha subunit | 2174 | 0.089 | 0.6190 | No |
| 11 | HIBCH | HIBCH Entrez, Source | 3-hydroxyisobutyryl-Coenzyme A hydrolase | 2392 | 0.083 | 0.6254 | No |
| 12 | PCCB | PCCB Entrez, Source | propionyl Coenzyme A carboxylase, beta polypeptide | 3417 | 0.058 | 0.5644 | No |
| 13 | ALDH7A1 | ALDH7A1 Entrez, Source | aldehyde dehydrogenase 7 family, member A1 | 3614 | 0.053 | 0.5644 | No |
| 14 | ACADM | ACADM Entrez, Source | acyl-Coenzyme A dehydrogenase, C-4 to C-12 straight chain | 4299 | 0.041 | 0.5245 | No |
| 15 | ALDH3A2 | ALDH3A2 Entrez, Source | aldehyde dehydrogenase 3 family, member A2 | 4819 | 0.032 | 0.4943 | No |
| 16 | ALDH1B1 | ALDH1B1 Entrez, Source | aldehyde dehydrogenase 1 family, member B1 | 5218 | 0.026 | 0.4716 | No |
| 17 | SUCLG1 | SUCLG1 Entrez, Source | succinate-CoA ligase, GDP-forming, alpha subunit | 5738 | 0.018 | 0.4377 | No |
| 18 | ALDH2 | ALDH2 Entrez, Source | aldehyde dehydrogenase 2 family (mitochondrial) | 6175 | 0.012 | 0.4082 | No |
| 19 | LDHC | LDHC Entrez, Source | lactate dehydrogenase C | 6640 | 0.005 | 0.3747 | No |
| 20 | SUCLG2 | SUCLG2 Entrez, Source | succinate-CoA ligase, GDP-forming, beta subunit | 7511 | -0.008 | 0.3115 | No |
| 21 | LDHA | LDHA Entrez, Source | lactate dehydrogenase A | 7676 | -0.010 | 0.3019 | No |
| 22 | LDHAL6B | LDHAL6B Entrez, Source | lactate dehydrogenase A-like 6B | 8335 | -0.020 | 0.2581 | No |
| 23 | ALDH3A1 | ALDH3A1 Entrez, Source | aldehyde dehydrogenase 3 family, memberA1 | 9824 | -0.048 | 0.1597 | No |
| 24 | ALDH9A1 | ALDH9A1 Entrez, Source | aldehyde dehydrogenase 9 family, member A1 | 10674 | -0.069 | 0.1150 | No |
| 25 | LDHB | LDHB Entrez, Source | lactate dehydrogenase B | 13034 | -0.309 | 0.0231 | No |