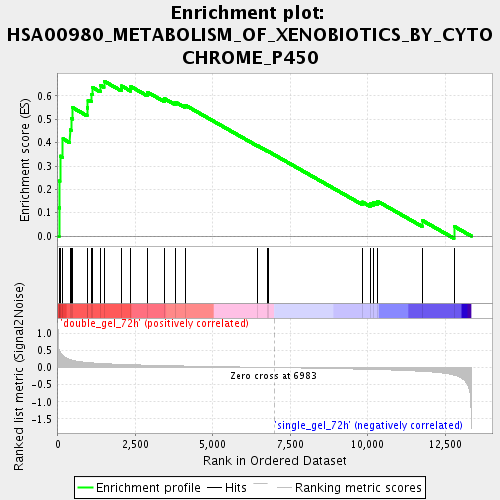
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_72h_versus_single_gel_72h.class.cls #double_gel_72h_versus_single_gel_72h_repos |
| Phenotype | class.cls#double_gel_72h_versus_single_gel_72h_repos |
| Upregulated in class | double_gel_72h |
| GeneSet | HSA00980_METABOLISM_OF_XENOBIOTICS_BY_CYTOCHROME_P450 |
| Enrichment Score (ES) | 0.6637842 |
| Normalized Enrichment Score (NES) | 2.0146735 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0022755903 |
| FWER p-Value | 0.1 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | GSTA2 | GSTA2 Entrez, Source | glutathione S-transferase A2 | 39 | 0.527 | 0.1210 | Yes |
| 2 | ADH7 | ADH7 Entrez, Source | alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide | 53 | 0.495 | 0.2363 | Yes |
| 3 | UGT2A1 | UGT2A1 Entrez, Source | UDP glucuronosyltransferase 2 family, polypeptide A1 | 68 | 0.458 | 0.3431 | Yes |
| 4 | ADHFE1 | ADHFE1 Entrez, Source | alcohol dehydrogenase, iron containing, 1 | 163 | 0.343 | 0.4166 | Yes |
| 5 | CYP1A2 | CYP1A2 Entrez, Source | cytochrome P450, family 1, subfamily A, polypeptide 2 | 390 | 0.235 | 0.4548 | Yes |
| 6 | GSTT2 | GSTT2 Entrez, Source | glutathione S-transferase theta 2 | 429 | 0.222 | 0.5041 | Yes |
| 7 | GSTK1 | GSTK1 Entrez, Source | glutathione S-transferase kappa 1 | 471 | 0.210 | 0.5503 | Yes |
| 8 | GSTM3 | GSTM3 Entrez, Source | glutathione S-transferase M3 (brain) | 943 | 0.149 | 0.5500 | Yes |
| 9 | GSTT1 | GSTT1 Entrez, Source | glutathione S-transferase theta 1 | 970 | 0.147 | 0.5828 | Yes |
| 10 | CYP1B1 | CYP1B1 Entrez, Source | cytochrome P450, family 1, subfamily B, polypeptide 1 | 1078 | 0.138 | 0.6073 | Yes |
| 11 | ADH1A | ADH1A Entrez, Source | alcohol dehydrogenase 1A (class I), alpha polypeptide | 1116 | 0.136 | 0.6364 | Yes |
| 12 | CYP2E1 | CYP2E1 Entrez, Source | cytochrome P450, family 2, subfamily E, polypeptide 1 | 1369 | 0.121 | 0.6459 | Yes |
| 13 | GSTA3 | GSTA3 Entrez, Source | glutathione S-transferase A3 | 1488 | 0.114 | 0.6638 | Yes |
| 14 | ADH6 | ADH6 Entrez, Source | alcohol dehydrogenase 6 (class V) | 2053 | 0.092 | 0.6431 | No |
| 15 | CYP1A1 | CYP1A1 Entrez, Source | cytochrome P450, family 1, subfamily A, polypeptide 1 | 2351 | 0.084 | 0.6405 | No |
| 16 | MGST1 | MGST1 Entrez, Source | microsomal glutathione S-transferase 1 | 2888 | 0.069 | 0.6166 | No |
| 17 | GSTO2 | GSTO2 Entrez, Source | glutathione S-transferase omega 2 | 3440 | 0.057 | 0.5886 | No |
| 18 | ADH4 | ADH4 Entrez, Source | alcohol dehydrogenase 4 (class II), pi polypeptide | 3800 | 0.050 | 0.5733 | No |
| 19 | EPHX1 | EPHX1 Entrez, Source | epoxide hydrolase 1, microsomal (xenobiotic) | 4104 | 0.045 | 0.5611 | No |
| 20 | GSTM1 | GSTM1 Entrez, Source | glutathione S-transferase M1 | 6456 | 0.007 | 0.3862 | No |
| 21 | CYP2C9 | CYP2C9 Entrez, Source | cytochrome P450, family 2, subfamily C, polypeptide 9 | 6753 | 0.003 | 0.3647 | No |
| 22 | GSTM5 | GSTM5 Entrez, Source | glutathione S-transferase M5 | 6794 | 0.003 | 0.3623 | No |
| 23 | ALDH3A1 | ALDH3A1 Entrez, Source | aldehyde dehydrogenase 3 family, memberA1 | 9824 | -0.048 | 0.1462 | No |
| 24 | GSTM2 | GSTM2 Entrez, Source | glutathione S-transferase M2 (muscle) | 10083 | -0.054 | 0.1394 | No |
| 25 | GSTM4 | GSTM4 Entrez, Source | glutathione S-transferase M4 | 10178 | -0.056 | 0.1455 | No |
| 26 | CYP3A4 | CYP3A4 Entrez, Source | cytochrome P450, family 3, subfamily A, polypeptide 4 | 10311 | -0.059 | 0.1496 | No |
| 27 | GSTA4 | GSTA4 Entrez, Source | glutathione S-transferase A4 | 11757 | -0.112 | 0.0673 | No |
| 28 | ALDH3B1 | ALDH3B1 Entrez, Source | aldehyde dehydrogenase 3 family, member B1 | 12784 | -0.220 | 0.0419 | No |