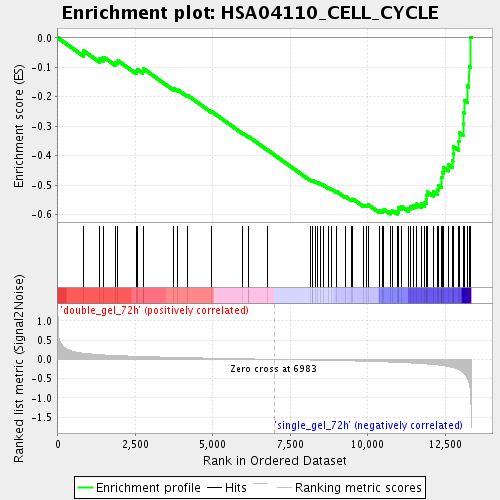
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_72h_versus_single_gel_72h.class.cls #double_gel_72h_versus_single_gel_72h_repos |
| Phenotype | class.cls#double_gel_72h_versus_single_gel_72h_repos |
| Upregulated in class | single_gel_72h |
| GeneSet | HSA04110_CELL_CYCLE |
| Enrichment Score (ES) | -0.5987888 |
| Normalized Enrichment Score (NES) | -1.9653172 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.009832042 |
| FWER p-Value | 0.262 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | GADD45G | GADD45G Entrez, Source | growth arrest and DNA-damage-inducible, gamma | 829 | 0.160 | -0.0441 | No |
| 2 | MAD2L2 | MAD2L2 Entrez, Source | MAD2 mitotic arrest deficient-like 2 (yeast) | 1355 | 0.121 | -0.0697 | No |
| 3 | CDC26 | CDC26 Entrez, Source | cell division cycle 26 | 1478 | 0.114 | -0.0657 | No |
| 4 | GADD45B | GADD45B Entrez, Source | growth arrest and DNA-damage-inducible, beta | 1856 | 0.099 | -0.0827 | No |
| 5 | CCNH | CCNH Entrez, Source | cyclin H | 1938 | 0.096 | -0.0778 | No |
| 6 | CDC25A | CDC25A Entrez, Source | cell division cycle 25A | 2527 | 0.079 | -0.1130 | No |
| 7 | CCNE1 | CCNE1 Entrez, Source | cyclin E1 | 2560 | 0.078 | -0.1064 | No |
| 8 | CDK6 | CDK6 Entrez, Source | cyclin-dependent kinase 6 | 2746 | 0.073 | -0.1120 | No |
| 9 | RB1 | RB1 Entrez, Source | retinoblastoma 1 (including osteosarcoma) | 2765 | 0.072 | -0.1051 | No |
| 10 | SMC1A | SMC1A Entrez, Source | structural maintenance of chromosomes 1A | 3744 | 0.051 | -0.1729 | No |
| 11 | RBL2 | RBL2 Entrez, Source | retinoblastoma-like 2 (p130) | 3874 | 0.049 | -0.1770 | No |
| 12 | ORC4L | ORC4L Entrez, Source | origin recognition complex, subunit 4-like (yeast) | 4185 | 0.043 | -0.1954 | No |
| 13 | ORC2L | ORC2L Entrez, Source | origin recognition complex, subunit 2-like (yeast) | 4948 | 0.030 | -0.2493 | No |
| 14 | PCNA | PCNA Entrez, Source | proliferating cell nuclear antigen | 5968 | 0.015 | -0.3244 | No |
| 15 | TP53 | TP53 Entrez, Source | tumor protein p53 (Li-Fraumeni syndrome) | 6158 | 0.012 | -0.3373 | No |
| 16 | CDC16 | CDC16 Entrez, Source | CDC16 cell division cycle 16 homolog (S. cerevisiae) | 6164 | 0.012 | -0.3363 | No |
| 17 | CDKN2A | CDKN2A Entrez, Source | cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4) | 6755 | 0.003 | -0.3804 | No |
| 18 | YWHAE | YWHAE Entrez, Source | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide | 8149 | -0.017 | -0.4834 | No |
| 19 | CDK2 | CDK2 Entrez, Source | cyclin-dependent kinase 2 | 8216 | -0.018 | -0.4862 | No |
| 20 | RBX1 | RBX1 Entrez, Source | ring-box 1 | 8219 | -0.018 | -0.4843 | No |
| 21 | CDKN1A | CDKN1A Entrez, Source | cyclin-dependent kinase inhibitor 1A (p21, Cip1) | 8319 | -0.020 | -0.4894 | No |
| 22 | CCNA1 | CCNA1 Entrez, Source | cyclin A1 | 8384 | -0.021 | -0.4918 | No |
| 23 | CDK7 | CDK7 Entrez, Source | cyclin-dependent kinase 7 (MO15 homolog, Xenopus laevis, cdk-activating kinase) | 8462 | -0.022 | -0.4950 | No |
| 24 | CREBBP | CREBBP Entrez, Source | CREB binding protein (Rubinstein-Taybi syndrome) | 8561 | -0.024 | -0.4996 | No |
| 25 | PLK1 | PLK1 Entrez, Source | polo-like kinase 1 (Drosophila) | 8736 | -0.027 | -0.5096 | No |
| 26 | YWHAB | YWHAB Entrez, Source | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide | 8834 | -0.029 | -0.5137 | No |
| 27 | BUB3 | BUB3 Entrez, Source | BUB3 budding uninhibited by benzimidazoles 3 homolog (yeast) | 8998 | -0.032 | -0.5222 | No |
| 28 | CDKN2C | CDKN2C Entrez, Source | cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4) | 9276 | -0.037 | -0.5388 | No |
| 29 | SMAD3 | SMAD3 Entrez, Source | SMAD, mothers against DPP homolog 3 (Drosophila) | 9459 | -0.041 | -0.5478 | No |
| 30 | ORC1L | ORC1L Entrez, Source | origin recognition complex, subunit 1-like (yeast) | 9519 | -0.042 | -0.5474 | No |
| 31 | CCND3 | CCND3 Entrez, Source | cyclin D3 | 9869 | -0.049 | -0.5681 | No |
| 32 | GADD45A | GADD45A Entrez, Source | growth arrest and DNA-damage-inducible, alpha | 9946 | -0.051 | -0.5679 | No |
| 33 | CDKN1C | CDKN1C Entrez, Source | cyclin-dependent kinase inhibitor 1C (p57, Kip2) | 10008 | -0.052 | -0.5665 | No |
| 34 | ORC3L | ORC3L Entrez, Source | origin recognition complex, subunit 3-like (yeast) | 10375 | -0.061 | -0.5871 | No |
| 35 | ANAPC4 | ANAPC4 Entrez, Source | anaphase promoting complex subunit 4 | 10467 | -0.063 | -0.5867 | No |
| 36 | CDC23 | CDC23 Entrez, Source | CDC23 (cell division cycle 23, yeast, homolog) | 10523 | -0.065 | -0.5833 | No |
| 37 | CDKN2B | CDKN2B Entrez, Source | cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4) | 10726 | -0.071 | -0.5905 | Yes |
| 38 | TGFB1 | TGFB1 Entrez, Source | transforming growth factor, beta 1 (Camurati-Engelmann disease) | 10788 | -0.072 | -0.5867 | Yes |
| 39 | CDKN2D | CDKN2D Entrez, Source | cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) | 10949 | -0.077 | -0.5899 | Yes |
| 40 | YWHAH | YWHAH Entrez, Source | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide | 10999 | -0.079 | -0.5846 | Yes |
| 41 | EP300 | EP300 Entrez, Source | E1A binding protein p300 | 11002 | -0.079 | -0.5757 | Yes |
| 42 | SMAD4 | SMAD4 Entrez, Source | SMAD, mothers against DPP homolog 4 (Drosophila) | 11091 | -0.082 | -0.5729 | Yes |
| 43 | HDAC2 | HDAC2 Entrez, Source | histone deacetylase 2 | 11317 | -0.091 | -0.5794 | Yes |
| 44 | YWHAG | YWHAG Entrez, Source | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide | 11367 | -0.093 | -0.5723 | Yes |
| 45 | GSK3B | GSK3B Entrez, Source | glycogen synthase kinase 3 beta | 11474 | -0.098 | -0.5690 | Yes |
| 46 | ORC6L | ORC6L Entrez, Source | origin recognition complex, subunit 6 like (yeast) | 11568 | -0.103 | -0.5642 | Yes |
| 47 | YWHAZ | YWHAZ Entrez, Source | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide | 11724 | -0.110 | -0.5632 | Yes |
| 48 | ANAPC2 | ANAPC2 Entrez, Source | anaphase promoting complex subunit 2 | 11844 | -0.116 | -0.5587 | Yes |
| 49 | CDKN1B | CDKN1B Entrez, Source | cyclin-dependent kinase inhibitor 1B (p27, Kip1) | 11886 | -0.118 | -0.5482 | Yes |
| 50 | CDC27 | CDC27 Entrez, Source | cell division cycle 27 | 11905 | -0.119 | -0.5359 | Yes |
| 51 | SMAD2 | SMAD2 Entrez, Source | SMAD, mothers against DPP homolog 2 (Drosophila) | 11923 | -0.120 | -0.5235 | Yes |
| 52 | TGFB2 | TGFB2 Entrez, Source | transforming growth factor, beta 2 | 12112 | -0.132 | -0.5224 | Yes |
| 53 | CDC20 | CDC20 Entrez, Source | CDC20 cell division cycle 20 homolog (S. cerevisiae) | 12256 | -0.144 | -0.5166 | Yes |
| 54 | CDK4 | CDK4 Entrez, Source | cyclin-dependent kinase 4 | 12272 | -0.146 | -0.5010 | Yes |
| 55 | E2F1 | E2F1 Entrez, Source | E2F transcription factor 1 | 12376 | -0.156 | -0.4908 | Yes |
| 56 | CDC25B | CDC25B Entrez, Source | cell division cycle 25B | 12377 | -0.156 | -0.4729 | Yes |
| 57 | MCM7 | MCM7 Entrez, Source | MCM7 minichromosome maintenance deficient 7 (S. cerevisiae) | 12413 | -0.160 | -0.4572 | Yes |
| 58 | CHEK2 | CHEK2 Entrez, Source | CHK2 checkpoint homolog (S. pombe) | 12445 | -0.163 | -0.4408 | Yes |
| 59 | YWHAQ | YWHAQ Entrez, Source | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide | 12601 | -0.184 | -0.4312 | Yes |
| 60 | CHEK1 | CHEK1 Entrez, Source | CHK1 checkpoint homolog (S. pombe) | 12725 | -0.209 | -0.4165 | Yes |
| 61 | PTTG1 | PTTG1 Entrez, Source | pituitary tumor-transforming 1 | 12752 | -0.214 | -0.3938 | Yes |
| 62 | BUB1B | BUB1B Entrez, Source | BUB1 budding uninhibited by benzimidazoles 1 homolog beta (yeast) | 12774 | -0.218 | -0.3704 | Yes |
| 63 | WEE1 | WEE1 Entrez, Source | WEE1 homolog (S. pombe) | 12929 | -0.262 | -0.3518 | Yes |
| 64 | CCNB2 | CCNB2 Entrez, Source | cyclin B2 | 12949 | -0.272 | -0.3220 | Yes |
| 65 | CCND2 | CCND2 Entrez, Source | cyclin D2 | 13080 | -0.340 | -0.2926 | Yes |
| 66 | CCND1 | CCND1 Entrez, Source | cyclin D1 | 13085 | -0.343 | -0.2535 | Yes |
| 67 | CCNB1 | CCNB1 Entrez, Source | cyclin B1 | 13131 | -0.385 | -0.2126 | Yes |
| 68 | TGFB3 | TGFB3 Entrez, Source | transforming growth factor, beta 3 | 13210 | -0.480 | -0.1632 | Yes |
| 69 | MCM6 | MCM6 Entrez, Source | MCM6 minichromosome maintenance deficient 6 (MIS5 homolog, S. pombe) (S. cerevisiae) | 13267 | -0.614 | -0.0968 | Yes |
| 70 | CCNA2 | CCNA2 Entrez, Source | cyclin A2 | 13314 | -0.890 | 0.0021 | Yes |