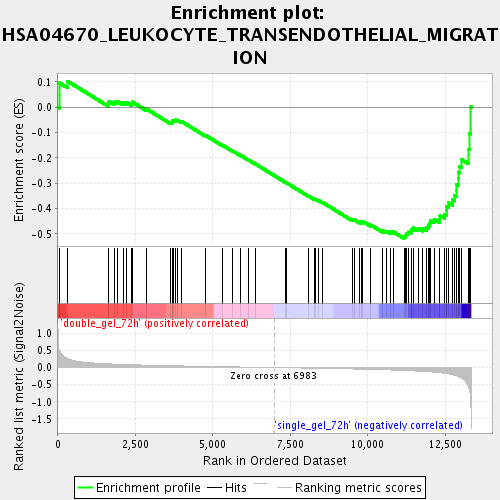
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_72h_versus_single_gel_72h.class.cls #double_gel_72h_versus_single_gel_72h_repos |
| Phenotype | class.cls#double_gel_72h_versus_single_gel_72h_repos |
| Upregulated in class | single_gel_72h |
| GeneSet | HSA04670_LEUKOCYTE_TRANSENDOTHELIAL_MIGRATION |
| Enrichment Score (ES) | -0.51787096 |
| Normalized Enrichment Score (NES) | -1.7007422 |
| Nominal p-value | 0.0034188034 |
| FDR q-value | 0.062815845 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | VCAM1 | VCAM1 Entrez, Source | vascular cell adhesion molecule 1 | 49 | 0.501 | 0.0461 | No |
| 2 | CXCL12 | CXCL12 Entrez, Source | chemokine (C-X-C motif) ligand 12 (stromal cell-derived factor 1) | 50 | 0.500 | 0.0958 | No |
| 3 | CLDN14 | CLDN14 Entrez, Source | claudin 14 | 306 | 0.265 | 0.1029 | No |
| 4 | VAV1 | VAV1 Entrez, Source | vav 1 oncogene | 1642 | 0.107 | 0.0129 | No |
| 5 | ICAM1 | ICAM1 Entrez, Source | intercellular adhesion molecule 1 (CD54), human rhinovirus receptor | 1644 | 0.107 | 0.0235 | No |
| 6 | MAPK14 | MAPK14 Entrez, Source | mitogen-activated protein kinase 14 | 1814 | 0.101 | 0.0208 | No |
| 7 | CLDN11 | CLDN11 Entrez, Source | claudin 11 (oligodendrocyte transmembrane protein) | 1925 | 0.096 | 0.0221 | No |
| 8 | NOX1 | NOX1 Entrez, Source | NADPH oxidase 1 | 2105 | 0.091 | 0.0176 | No |
| 9 | ARHGAP5 | ARHGAP5 Entrez, Source | Rho GTPase activating protein 5 | 2217 | 0.088 | 0.0180 | No |
| 10 | CLDN8 | CLDN8 Entrez, Source | claudin 8 | 2374 | 0.083 | 0.0145 | No |
| 11 | PRKCB1 | PRKCB1 Entrez, Source | protein kinase C, beta 1 | 2401 | 0.082 | 0.0207 | No |
| 12 | RAPGEF3 | RAPGEF3 Entrez, Source | Rap guanine nucleotide exchange factor (GEF) 3 | 2860 | 0.070 | -0.0069 | No |
| 13 | CLDN3 | CLDN3 Entrez, Source | claudin 3 | 3644 | 0.053 | -0.0606 | No |
| 14 | MYL2 | MYL2 Entrez, Source | myosin, light chain 2, regulatory, cardiac, slow | 3684 | 0.052 | -0.0583 | No |
| 15 | ITGAM | ITGAM Entrez, Source | integrin, alpha M (complement component 3 receptor 3 subunit) | 3688 | 0.052 | -0.0534 | No |
| 16 | NCF1 | NCF1 Entrez, Source | neutrophil cytosolic factor 1, (chronic granulomatous disease, autosomal 1) | 3743 | 0.051 | -0.0524 | No |
| 17 | RAC2 | RAC2 Entrez, Source | ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) | 3781 | 0.050 | -0.0502 | No |
| 18 | MMP9 | MMP9 Entrez, Source | matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) | 3861 | 0.049 | -0.0513 | No |
| 19 | PRKCA | PRKCA Entrez, Source | protein kinase C, alpha | 3999 | 0.046 | -0.0570 | No |
| 20 | PIK3R1 | PIK3R1 Entrez, Source | phosphoinositide-3-kinase, regulatory subunit 1 (p85 alpha) | 4760 | 0.033 | -0.1110 | No |
| 21 | CYBB | CYBB Entrez, Source | cytochrome b-245, beta polypeptide (chronic granulomatous disease) | 5312 | 0.024 | -0.1501 | No |
| 22 | SIPA1 | SIPA1 Entrez, Source | signal-induced proliferation-associated gene 1 | 5630 | 0.020 | -0.1720 | No |
| 23 | RAP1A | RAP1A Entrez, Source | RAP1A, member of RAS oncogene family | 5896 | 0.016 | -0.1904 | No |
| 24 | JAM2 | JAM2 Entrez, Source | junctional adhesion molecule 2 | 6160 | 0.012 | -0.2090 | No |
| 25 | PIK3R2 | PIK3R2 Entrez, Source | phosphoinositide-3-kinase, regulatory subunit 2 (p85 beta) | 6361 | 0.009 | -0.2232 | No |
| 26 | TXK | TXK Entrez, Source | TXK tyrosine kinase | 7344 | -0.005 | -0.2967 | No |
| 27 | CLDN16 | CLDN16 Entrez, Source | claudin 16 | 7371 | -0.006 | -0.2981 | No |
| 28 | PTPN11 | PTPN11 Entrez, Source | protein tyrosine phosphatase, non-receptor type 11 (Noonan syndrome 1) | 8096 | -0.016 | -0.3511 | No |
| 29 | ITGB2 | ITGB2 Entrez, Source | integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) | 8281 | -0.020 | -0.3630 | No |
| 30 | CTNNB1 | CTNNB1 Entrez, Source | catenin (cadherin-associated protein), beta 1, 88kDa | 8312 | -0.020 | -0.3633 | No |
| 31 | PIK3CB | PIK3CB Entrez, Source | phosphoinositide-3-kinase, catalytic, beta polypeptide | 8414 | -0.022 | -0.3687 | No |
| 32 | MAPK13 | MAPK13 Entrez, Source | mitogen-activated protein kinase 13 | 8524 | -0.023 | -0.3746 | No |
| 33 | GNAI3 | GNAI3 Entrez, Source | guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 3 | 9496 | -0.042 | -0.4436 | No |
| 34 | CDC42 | CDC42 Entrez, Source | cell division cycle 42 (GTP binding protein, 25kDa) | 9556 | -0.043 | -0.4438 | No |
| 35 | CXCR4 | CXCR4 Entrez, Source | chemokine (C-X-C motif) receptor 4 | 9737 | -0.046 | -0.4528 | No |
| 36 | RASSF5 | RASSF5 Entrez, Source | Ras association (RalGDS/AF-6) domain family 5 | 9803 | -0.048 | -0.4529 | No |
| 37 | PLCG2 | PLCG2 Entrez, Source | phospholipase C, gamma 2 (phosphatidylinositol-specific) | 9827 | -0.048 | -0.4498 | No |
| 38 | CLDN1 | CLDN1 Entrez, Source | claudin 1 | 10094 | -0.054 | -0.4645 | No |
| 39 | CLDN4 | CLDN4 Entrez, Source | claudin 4 | 10487 | -0.064 | -0.4877 | No |
| 40 | CLDN5 | CLDN5 Entrez, Source | claudin 5 (transmembrane protein deleted in velocardiofacial syndrome) | 10600 | -0.067 | -0.4895 | No |
| 41 | RAC1 | RAC1 Entrez, Source | ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) | 10733 | -0.071 | -0.4924 | No |
| 42 | RHOH | RHOH Entrez, Source | ras homolog gene family, member H | 10818 | -0.073 | -0.4915 | No |
| 43 | THY1 | THY1 Entrez, Source | Thy-1 cell surface antigen | 11169 | -0.084 | -0.5095 | Yes |
| 44 | ROCK1 | ROCK1 Entrez, Source | Rho-associated, coiled-coil containing protein kinase 1 | 11231 | -0.087 | -0.5054 | Yes |
| 45 | CLDN19 | CLDN19 Entrez, Source | claudin 19 | 11250 | -0.088 | -0.4980 | Yes |
| 46 | F11R | F11R Entrez, Source | F11 receptor | 11299 | -0.090 | -0.4927 | Yes |
| 47 | ITGB1 | ITGB1 Entrez, Source | integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) | 11396 | -0.095 | -0.4905 | Yes |
| 48 | MLLT4 | MLLT4 Entrez, Source | myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 4 | 11397 | -0.095 | -0.4811 | Yes |
| 49 | RHOA | RHOA Entrez, Source | ras homolog gene family, member A | 11466 | -0.098 | -0.4764 | Yes |
| 50 | PIK3R3 | PIK3R3 Entrez, Source | phosphoinositide-3-kinase, regulatory subunit 3 (p55, gamma) | 11622 | -0.106 | -0.4776 | Yes |
| 51 | ITGAL | ITGAL Entrez, Source | integrin, alpha L (antigen CD11A (p180), lymphocyte function-associated antigen 1; alpha polypeptide) | 11774 | -0.112 | -0.4778 | Yes |
| 52 | PTK2 | PTK2 Entrez, Source | PTK2 protein tyrosine kinase 2 | 11904 | -0.119 | -0.4758 | Yes |
| 53 | ACTN3 | ACTN3 Entrez, Source | actinin, alpha 3 | 11968 | -0.123 | -0.4683 | Yes |
| 54 | RAPGEF4 | RAPGEF4 Entrez, Source | Rap guanine nucleotide exchange factor (GEF) 4 | 11990 | -0.124 | -0.4575 | Yes |
| 55 | BCAR1 | BCAR1 Entrez, Source | breast cancer anti-estrogen resistance 1 | 12011 | -0.126 | -0.4465 | Yes |
| 56 | RAP1B | RAP1B Entrez, Source | RAP1B, member of RAS oncogene family | 12148 | -0.134 | -0.4434 | Yes |
| 57 | CD99 | CD99 Entrez, Source | CD99 molecule | 12323 | -0.150 | -0.4416 | Yes |
| 58 | MAPK12 | MAPK12 Entrez, Source | mitogen-activated protein kinase 12 | 12330 | -0.151 | -0.4271 | Yes |
| 59 | GNAI2 | GNAI2 Entrez, Source | guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2 | 12486 | -0.168 | -0.4221 | Yes |
| 60 | PIK3CA | PIK3CA Entrez, Source | phosphoinositide-3-kinase, catalytic, alpha polypeptide | 12536 | -0.176 | -0.4082 | Yes |
| 61 | CLDN7 | CLDN7 Entrez, Source | claudin 7 | 12546 | -0.178 | -0.3912 | Yes |
| 62 | MMP2 | MMP2 Entrez, Source | matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase) | 12615 | -0.187 | -0.3778 | Yes |
| 63 | PXN | PXN Entrez, Source | paxillin | 12740 | -0.211 | -0.3661 | Yes |
| 64 | PLCG1 | PLCG1 Entrez, Source | phospholipase C, gamma 1 | 12798 | -0.224 | -0.3482 | Yes |
| 65 | ACTN4 | ACTN4 Entrez, Source | actinin, alpha 4 | 12871 | -0.243 | -0.3294 | Yes |
| 66 | PTK2B | PTK2B Entrez, Source | PTK2B protein tyrosine kinase 2 beta | 12874 | -0.243 | -0.3054 | Yes |
| 67 | CYBA | CYBA Entrez, Source | cytochrome b-245, alpha polypeptide | 12921 | -0.260 | -0.2831 | Yes |
| 68 | JAM3 | JAM3 Entrez, Source | junctional adhesion molecule 3 | 12925 | -0.260 | -0.2574 | Yes |
| 69 | ESAM | ESAM Entrez, Source | endothelial cell adhesion molecule | 12960 | -0.276 | -0.2325 | Yes |
| 70 | ROCK2 | ROCK2 Entrez, Source | Rho-associated, coiled-coil containing protein kinase 2 | 13035 | -0.310 | -0.2072 | Yes |
| 71 | GNAI1 | GNAI1 Entrez, Source | guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 | 13262 | -0.580 | -0.1666 | Yes |
| 72 | ACTN1 | ACTN1 Entrez, Source | actinin, alpha 1 | 13279 | -0.655 | -0.1027 | Yes |
| 73 | MSN | MSN Entrez, Source | moesin | 13329 | -1.080 | 0.0010 | Yes |