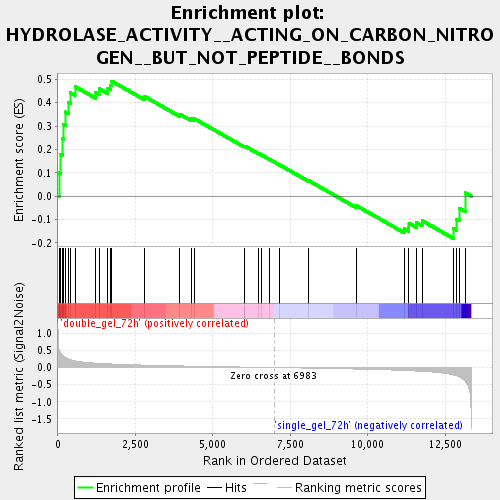
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_72h_versus_single_gel_72h.class.cls #double_gel_72h_versus_single_gel_72h_repos |
| Phenotype | class.cls#double_gel_72h_versus_single_gel_72h_repos |
| Upregulated in class | double_gel_72h |
| GeneSet | HYDROLASE_ACTIVITY__ACTING_ON_CARBON_NITROGEN__BUT_NOT_PEPTIDE__BONDS |
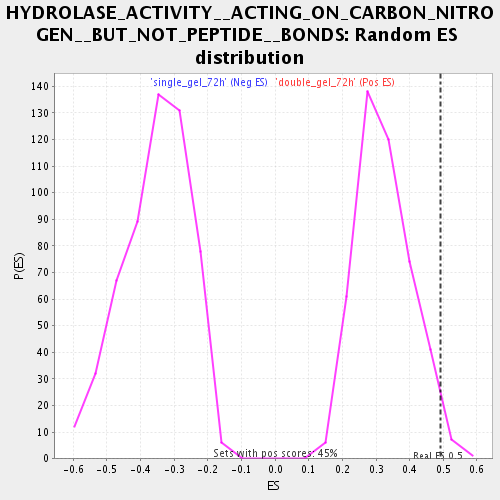
| Enrichment Score (ES) | 0.49238634 |
| Normalized Enrichment Score (NES) | 1.5218197 |
| Nominal p-value | 0.017857144 |
| FDR q-value | 0.109398514 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | APOBEC1 | APOBEC1 Entrez, Source | apolipoprotein B mRNA editing enzyme, catalytic polypeptide 1 | 40 | 0.526 | 0.1002 | Yes |
| 2 | DPYS | DPYS Entrez, Source | dihydropyrimidinase | 90 | 0.425 | 0.1800 | Yes |
| 3 | ARG1 | ARG1 Entrez, Source | arginase, liver | 145 | 0.358 | 0.2462 | Yes |
| 4 | UPB1 | UPB1 Entrez, Source | ureidopropionase, beta | 177 | 0.330 | 0.3086 | Yes |
| 5 | GCH1 | GCH1 Entrez, Source | GTP cyclohydrolase 1 (dopa-responsive dystonia) | 243 | 0.289 | 0.3604 | Yes |
| 6 | GLS2 | GLS2 Entrez, Source | glutaminase 2 (liver, mitochondrial) | 341 | 0.252 | 0.4024 | Yes |
| 7 | MTHFD1 | MTHFD1 Entrez, Source | methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase | 405 | 0.229 | 0.4426 | Yes |
| 8 | FAAH | FAAH Entrez, Source | fatty acid amide hydrolase | 553 | 0.192 | 0.4692 | Yes |
| 9 | ADA | ADA Entrez, Source | adenosine deaminase | 1219 | 0.129 | 0.4445 | Yes |
| 10 | ACY1 | ACY1 Entrez, Source | aminoacylase 1 | 1335 | 0.122 | 0.4598 | Yes |
| 11 | ASRGL1 | ASRGL1 Entrez, Source | asparaginase like 1 | 1612 | 0.108 | 0.4603 | Yes |
| 12 | GDA | GDA Entrez, Source | guanine deaminase | 1689 | 0.105 | 0.4753 | Yes |
| 13 | ASPA | ASPA Entrez, Source | aspartoacylase (Canavan disease) | 1733 | 0.104 | 0.4924 | Yes |
| 14 | PADI3 | PADI3 Entrez, Source | peptidyl arginine deiminase, type III | 2797 | 0.072 | 0.4266 | No |
| 15 | HDAC10 | HDAC10 Entrez, Source | histone deacetylase 10 | 3923 | 0.048 | 0.3514 | No |
| 16 | ADARB2 | ADARB2 Entrez, Source | adenosine deaminase, RNA-specific, B2 (RED2 homolog rat) | 4297 | 0.041 | 0.3315 | No |
| 17 | ACR | ACR Entrez, Source | acrosin | 4402 | 0.039 | 0.3314 | No |
| 18 | PADI4 | PADI4 Entrez, Source | peptidyl arginine deiminase, type IV | 6014 | 0.014 | 0.2131 | No |
| 19 | DPYSL2 | DPYSL2 Entrez, Source | dihydropyrimidinase-like 2 | 6036 | 0.014 | 0.2142 | No |
| 20 | DARS | DARS Entrez, Source | aspartyl-tRNA synthetase | 6478 | 0.007 | 0.1824 | No |
| 21 | ARG2 | ARG2 Entrez, Source | arginase, type II | 6580 | 0.006 | 0.1759 | No |
| 22 | HDAC3 | HDAC3 Entrez, Source | histone deacetylase 3 | 6844 | 0.002 | 0.1565 | No |
| 23 | NIT1 | NIT1 Entrez, Source | nitrilase 1 | 7155 | -0.002 | 0.1337 | No |
| 24 | AMPD1 | AMPD1 Entrez, Source | adenosine monophosphate deaminase 1 (isoform M) | 8100 | -0.016 | 0.0660 | No |
| 25 | ATIC | ATIC Entrez, Source | 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase | 9649 | -0.045 | -0.0416 | No |
| 26 | HDAC6 | HDAC6 Entrez, Source | histone deacetylase 6 | 11173 | -0.085 | -0.1395 | No |
| 27 | HDAC2 | HDAC2 Entrez, Source | histone deacetylase 2 | 11317 | -0.091 | -0.1323 | No |
| 28 | HDAC5 | HDAC5 Entrez, Source | histone deacetylase 5 | 11330 | -0.092 | -0.1152 | No |
| 29 | SIRT2 | SIRT2 Entrez, Source | sirtuin (silent mating type information regulation 2 homolog) 2 (S. cerevisiae) | 11576 | -0.103 | -0.1134 | No |
| 30 | AGA | AGA Entrez, Source | aspartylglucosaminidase | 11755 | -0.111 | -0.1049 | No |
| 31 | CRMP1 | CRMP1 Entrez, Source | collapsin response mediator protein 1 | 12762 | -0.215 | -0.1383 | No |
| 32 | HDAC7A | HDAC7A Entrez, Source | histone deacetylase 7A | 12873 | -0.243 | -0.0989 | No |
| 33 | AMPD3 | AMPD3 Entrez, Source | adenosine monophosphate deaminase (isoform E) | 12958 | -0.275 | -0.0513 | No |
| 34 | DPYSL3 | DPYSL3 Entrez, Source | dihydropyrimidinase-like 3 | 13155 | -0.408 | 0.0141 | No |