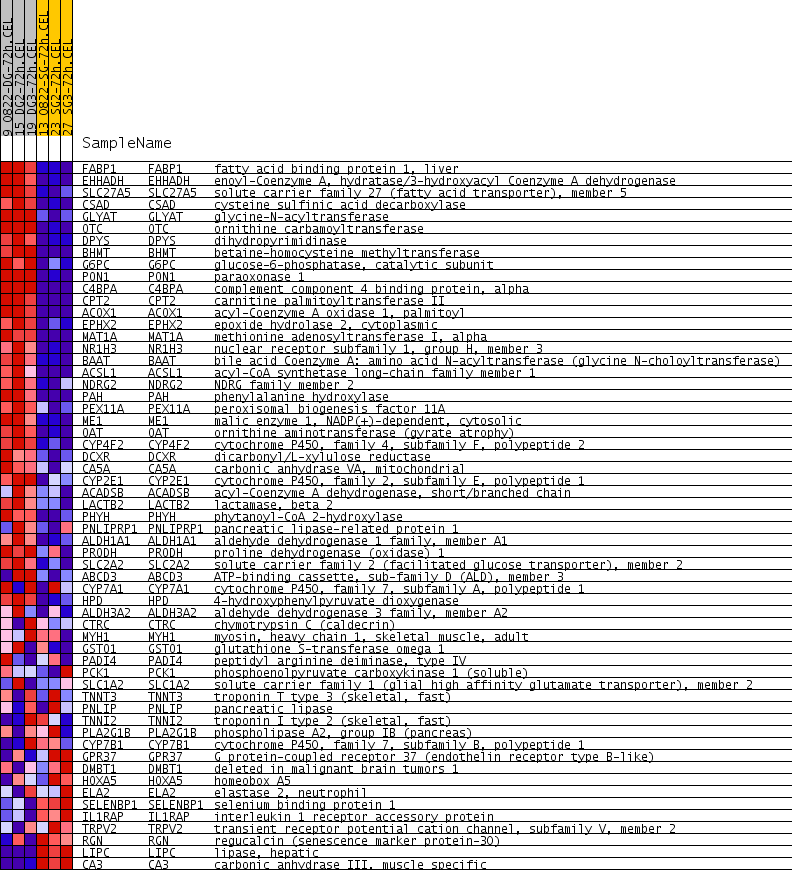
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_72h_versus_single_gel_72h.class.cls #double_gel_72h_versus_single_gel_72h_repos |
| Phenotype | class.cls#double_gel_72h_versus_single_gel_72h_repos |
| Upregulated in class | double_gel_72h |
| GeneSet | LEE_DENA_DN |
| Enrichment Score (ES) | 0.63831884 |
| Normalized Enrichment Score (NES) | 2.1712618 |
| Nominal p-value | 0.0 |
| FDR q-value | 2.1970617E-4 |
| FWER p-Value | 0.0070 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | FABP1 | FABP1 Entrez, Source | fatty acid binding protein 1, liver | 2 | 1.112 | 0.0849 | Yes |
| 2 | EHHADH | EHHADH Entrez, Source | enoyl-Coenzyme A, hydratase/3-hydroxyacyl Coenzyme A dehydrogenase | 6 | 0.908 | 0.1542 | Yes |
| 3 | SLC27A5 | SLC27A5 Entrez, Source | solute carrier family 27 (fatty acid transporter), member 5 | 9 | 0.779 | 0.2136 | Yes |
| 4 | CSAD | CSAD Entrez, Source | cysteine sulfinic acid decarboxylase | 23 | 0.595 | 0.2581 | Yes |
| 5 | GLYAT | GLYAT Entrez, Source | glycine-N-acyltransferase | 33 | 0.554 | 0.2999 | Yes |
| 6 | OTC | OTC Entrez, Source | ornithine carbamoyltransferase | 44 | 0.506 | 0.3378 | Yes |
| 7 | DPYS | DPYS Entrez, Source | dihydropyrimidinase | 90 | 0.425 | 0.3670 | Yes |
| 8 | BHMT | BHMT Entrez, Source | betaine-homocysteine methyltransferase | 106 | 0.415 | 0.3975 | Yes |
| 9 | G6PC | G6PC Entrez, Source | glucose-6-phosphatase, catalytic subunit | 129 | 0.383 | 0.4252 | Yes |
| 10 | PON1 | PON1 Entrez, Source | paraoxonase 1 | 159 | 0.345 | 0.4494 | Yes |
| 11 | C4BPA | C4BPA Entrez, Source | complement component 4 binding protein, alpha | 176 | 0.331 | 0.4735 | Yes |
| 12 | CPT2 | CPT2 Entrez, Source | carnitine palmitoyltransferase II | 207 | 0.307 | 0.4947 | Yes |
| 13 | ACOX1 | ACOX1 Entrez, Source | acyl-Coenzyme A oxidase 1, palmitoyl | 266 | 0.281 | 0.5119 | Yes |
| 14 | EPHX2 | EPHX2 Entrez, Source | epoxide hydrolase 2, cytoplasmic | 282 | 0.274 | 0.5317 | Yes |
| 15 | MAT1A | MAT1A Entrez, Source | methionine adenosyltransferase I, alpha | 315 | 0.262 | 0.5493 | Yes |
| 16 | NR1H3 | NR1H3 Entrez, Source | nuclear receptor subfamily 1, group H, member 3 | 583 | 0.188 | 0.5436 | Yes |
| 17 | BAAT | BAAT Entrez, Source | bile acid Coenzyme A: amino acid N-acyltransferase (glycine N-choloyltransferase) | 597 | 0.186 | 0.5568 | Yes |
| 18 | ACSL1 | ACSL1 Entrez, Source | acyl-CoA synthetase long-chain family member 1 | 609 | 0.185 | 0.5702 | Yes |
| 19 | NDRG2 | NDRG2 Entrez, Source | NDRG family member 2 | 610 | 0.185 | 0.5843 | Yes |
| 20 | PAH | PAH Entrez, Source | phenylalanine hydroxylase | 628 | 0.182 | 0.5970 | Yes |
| 21 | PEX11A | PEX11A Entrez, Source | peroxisomal biogenesis factor 11A | 631 | 0.182 | 0.6107 | Yes |
| 22 | ME1 | ME1 Entrez, Source | malic enzyme 1, NADP(+)-dependent, cytosolic | 800 | 0.163 | 0.6105 | Yes |
| 23 | OAT | OAT Entrez, Source | ornithine aminotransferase (gyrate atrophy) | 893 | 0.154 | 0.6154 | Yes |
| 24 | CYP4F2 | CYP4F2 Entrez, Source | cytochrome P450, family 4, subfamily F, polypeptide 2 | 933 | 0.150 | 0.6239 | Yes |
| 25 | DCXR | DCXR Entrez, Source | dicarbonyl/L-xylulose reductase | 1010 | 0.144 | 0.6292 | Yes |
| 26 | CA5A | CA5A Entrez, Source | carbonic anhydrase VA, mitochondrial | 1034 | 0.142 | 0.6383 | Yes |
| 27 | CYP2E1 | CYP2E1 Entrez, Source | cytochrome P450, family 2, subfamily E, polypeptide 1 | 1369 | 0.121 | 0.6224 | No |
| 28 | ACADSB | ACADSB Entrez, Source | acyl-Coenzyme A dehydrogenase, short/branched chain | 1480 | 0.114 | 0.6229 | No |
| 29 | LACTB2 | LACTB2 Entrez, Source | lactamase, beta 2 | 1593 | 0.109 | 0.6228 | No |
| 30 | PHYH | PHYH Entrez, Source | phytanoyl-CoA 2-hydroxylase | 2036 | 0.093 | 0.5966 | No |
| 31 | PNLIPRP1 | PNLIPRP1 Entrez, Source | pancreatic lipase-related protein 1 | 2104 | 0.091 | 0.5985 | No |
| 32 | ALDH1A1 | ALDH1A1 Entrez, Source | aldehyde dehydrogenase 1 family, member A1 | 2239 | 0.087 | 0.5950 | No |
| 33 | PRODH | PRODH Entrez, Source | proline dehydrogenase (oxidase) 1 | 2331 | 0.084 | 0.5946 | No |
| 34 | SLC2A2 | SLC2A2 Entrez, Source | solute carrier family 2 (facilitated glucose transporter), member 2 | 2459 | 0.081 | 0.5913 | No |
| 35 | ABCD3 | ABCD3 Entrez, Source | ATP-binding cassette, sub-family D (ALD), member 3 | 3314 | 0.060 | 0.5315 | No |
| 36 | CYP7A1 | CYP7A1 Entrez, Source | cytochrome P450, family 7, subfamily A, polypeptide 1 | 4020 | 0.046 | 0.4820 | No |
| 37 | HPD | HPD Entrez, Source | 4-hydroxyphenylpyruvate dioxygenase | 4383 | 0.040 | 0.4578 | No |
| 38 | ALDH3A2 | ALDH3A2 Entrez, Source | aldehyde dehydrogenase 3 family, member A2 | 4819 | 0.032 | 0.4275 | No |
| 39 | CTRC | CTRC Entrez, Source | chymotrypsin C (caldecrin) | 4883 | 0.031 | 0.4251 | No |
| 40 | MYH1 | MYH1 Entrez, Source | myosin, heavy chain 1, skeletal muscle, adult | 5171 | 0.027 | 0.4056 | No |
| 41 | GSTO1 | GSTO1 Entrez, Source | glutathione S-transferase omega 1 | 5429 | 0.023 | 0.3880 | No |
| 42 | PADI4 | PADI4 Entrez, Source | peptidyl arginine deiminase, type IV | 6014 | 0.014 | 0.3451 | No |
| 43 | PCK1 | PCK1 Entrez, Source | phosphoenolpyruvate carboxykinase 1 (soluble) | 6026 | 0.014 | 0.3453 | No |
| 44 | SLC1A2 | SLC1A2 Entrez, Source | solute carrier family 1 (glial high affinity glutamate transporter), member 2 | 6115 | 0.013 | 0.3396 | No |
| 45 | TNNT3 | TNNT3 Entrez, Source | troponin T type 3 (skeletal, fast) | 6538 | 0.006 | 0.3083 | No |
| 46 | PNLIP | PNLIP Entrez, Source | pancreatic lipase | 6618 | 0.005 | 0.3027 | No |
| 47 | TNNI2 | TNNI2 Entrez, Source | troponin I type 2 (skeletal, fast) | 7460 | -0.007 | 0.2400 | No |
| 48 | PLA2G1B | PLA2G1B Entrez, Source | phospholipase A2, group IB (pancreas) | 8831 | -0.029 | 0.1390 | No |
| 49 | CYP7B1 | CYP7B1 Entrez, Source | cytochrome P450, family 7, subfamily B, polypeptide 1 | 9114 | -0.034 | 0.1204 | No |
| 50 | GPR37 | GPR37 Entrez, Source | G protein-coupled receptor 37 (endothelin receptor type B-like) | 9392 | -0.040 | 0.1026 | No |
| 51 | DMBT1 | DMBT1 Entrez, Source | deleted in malignant brain tumors 1 | 9974 | -0.052 | 0.0628 | No |
| 52 | HOXA5 | HOXA5 Entrez, Source | homeobox A5 | 10111 | -0.055 | 0.0567 | No |
| 53 | ELA2 | ELA2 Entrez, Source | elastase 2, neutrophil | 10427 | -0.062 | 0.0378 | No |
| 54 | SELENBP1 | SELENBP1 Entrez, Source | selenium binding protein 1 | 11162 | -0.084 | -0.0111 | No |
| 55 | IL1RAP | IL1RAP Entrez, Source | interleukin 1 receptor accessory protein | 11842 | -0.116 | -0.0533 | No |
| 56 | TRPV2 | TRPV2 Entrez, Source | transient receptor potential cation channel, subfamily V, member 2 | 11876 | -0.118 | -0.0468 | No |
| 57 | RGN | RGN Entrez, Source | regucalcin (senescence marker protein-30) | 12427 | -0.161 | -0.0758 | No |
| 58 | LIPC | LIPC Entrez, Source | lipase, hepatic | 13275 | -0.650 | -0.0899 | No |
| 59 | CA3 | CA3 Entrez, Source | carbonic anhydrase III, muscle specific | 13334 | -1.240 | 0.0006 | No |