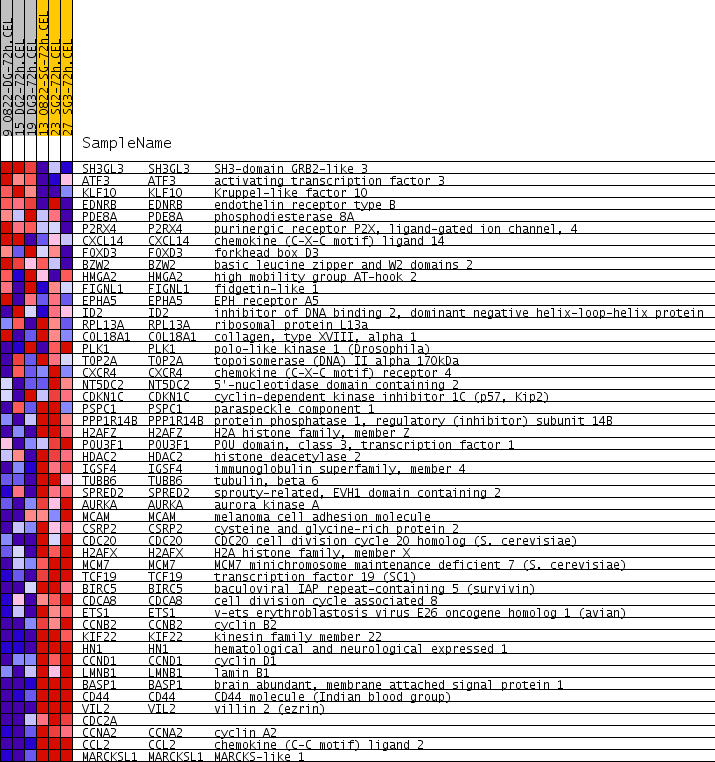
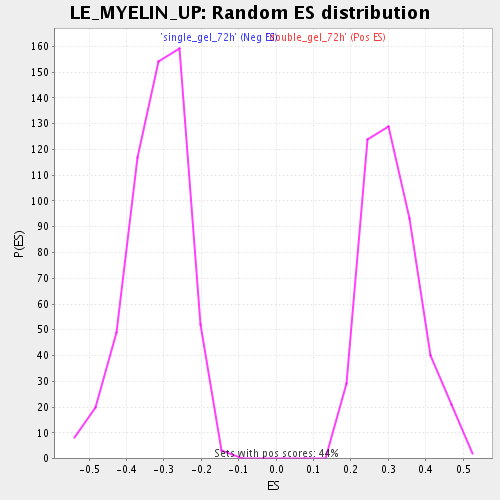
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_72h_versus_single_gel_72h.class.cls #double_gel_72h_versus_single_gel_72h_repos |
| Phenotype | class.cls#double_gel_72h_versus_single_gel_72h_repos |
| Upregulated in class | single_gel_72h |
| GeneSet | LE_MYELIN_UP |
| Enrichment Score (ES) | -0.7247998 |
| Normalized Enrichment Score (NES) | -2.2755187 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | SH3GL3 | SH3GL3 Entrez, Source | SH3-domain GRB2-like 3 | 397 | 0.234 | -0.0063 | No |
| 2 | ATF3 | ATF3 Entrez, Source | activating transcription factor 3 | 1413 | 0.118 | -0.0707 | No |
| 3 | KLF10 | KLF10 Entrez, Source | Kruppel-like factor 10 | 1693 | 0.105 | -0.0811 | No |
| 4 | EDNRB | EDNRB Entrez, Source | endothelin receptor type B | 1747 | 0.103 | -0.0746 | No |
| 5 | PDE8A | PDE8A Entrez, Source | phosphodiesterase 8A | 2516 | 0.079 | -0.1244 | No |
| 6 | P2RX4 | P2RX4 Entrez, Source | purinergic receptor P2X, ligand-gated ion channel, 4 | 3178 | 0.063 | -0.1678 | No |
| 7 | CXCL14 | CXCL14 Entrez, Source | chemokine (C-X-C motif) ligand 14 | 3372 | 0.059 | -0.1764 | No |
| 8 | FOXD3 | FOXD3 Entrez, Source | forkhead box D3 | 3412 | 0.058 | -0.1735 | No |
| 9 | BZW2 | BZW2 Entrez, Source | basic leucine zipper and W2 domains 2 | 4979 | 0.029 | -0.2883 | No |
| 10 | HMGA2 | HMGA2 Entrez, Source | high mobility group AT-hook 2 | 5052 | 0.028 | -0.2908 | No |
| 11 | FIGNL1 | FIGNL1 Entrez, Source | fidgetin-like 1 | 5215 | 0.026 | -0.3004 | No |
| 12 | EPHA5 | EPHA5 Entrez, Source | EPH receptor A5 | 5434 | 0.023 | -0.3145 | No |
| 13 | ID2 | ID2 Entrez, Source | inhibitor of DNA binding 2, dominant negative helix-loop-helix protein | 6773 | 0.003 | -0.4149 | No |
| 14 | RPL13A | RPL13A Entrez, Source | ribosomal protein L13a | 7446 | -0.007 | -0.4648 | No |
| 15 | COL18A1 | COL18A1 Entrez, Source | collagen, type XVIII, alpha 1 | 8284 | -0.020 | -0.5257 | No |
| 16 | PLK1 | PLK1 Entrez, Source | polo-like kinase 1 (Drosophila) | 8736 | -0.027 | -0.5569 | No |
| 17 | TOP2A | TOP2A Entrez, Source | topoisomerase (DNA) II alpha 170kDa | 9129 | -0.035 | -0.5829 | No |
| 18 | CXCR4 | CXCR4 Entrez, Source | chemokine (C-X-C motif) receptor 4 | 9737 | -0.046 | -0.6239 | No |
| 19 | NT5DC2 | NT5DC2 Entrez, Source | 5'-nucleotidase domain containing 2 | 9788 | -0.048 | -0.6228 | No |
| 20 | CDKN1C | CDKN1C Entrez, Source | cyclin-dependent kinase inhibitor 1C (p57, Kip2) | 10008 | -0.052 | -0.6340 | No |
| 21 | PSPC1 | PSPC1 Entrez, Source | paraspeckle component 1 | 10148 | -0.056 | -0.6389 | No |
| 22 | PPP1R14B | PPP1R14B Entrez, Source | protein phosphatase 1, regulatory (inhibitor) subunit 14B | 10466 | -0.063 | -0.6563 | No |
| 23 | H2AFZ | H2AFZ Entrez, Source | H2A histone family, member Z | 11071 | -0.081 | -0.6936 | No |
| 24 | POU3F1 | POU3F1 Entrez, Source | POU domain, class 3, transcription factor 1 | 11131 | -0.083 | -0.6897 | No |
| 25 | HDAC2 | HDAC2 Entrez, Source | histone deacetylase 2 | 11317 | -0.091 | -0.6943 | No |
| 26 | IGSF4 | IGSF4 Entrez, Source | immunoglobulin superfamily, member 4 | 11649 | -0.107 | -0.7085 | No |
| 27 | TUBB6 | TUBB6 Entrez, Source | tubulin, beta 6 | 11867 | -0.117 | -0.7129 | Yes |
| 28 | SPRED2 | SPRED2 Entrez, Source | sprouty-related, EVH1 domain containing 2 | 11916 | -0.119 | -0.7045 | Yes |
| 29 | AURKA | AURKA Entrez, Source | aurora kinase A | 11917 | -0.119 | -0.6925 | Yes |
| 30 | MCAM | MCAM Entrez, Source | melanoma cell adhesion molecule | 11953 | -0.122 | -0.6828 | Yes |
| 31 | CSRP2 | CSRP2 Entrez, Source | cysteine and glycine-rich protein 2 | 12164 | -0.135 | -0.6849 | Yes |
| 32 | CDC20 | CDC20 Entrez, Source | CDC20 cell division cycle 20 homolog (S. cerevisiae) | 12256 | -0.144 | -0.6772 | Yes |
| 33 | H2AFX | H2AFX Entrez, Source | H2A histone family, member X | 12313 | -0.149 | -0.6663 | Yes |
| 34 | MCM7 | MCM7 Entrez, Source | MCM7 minichromosome maintenance deficient 7 (S. cerevisiae) | 12413 | -0.160 | -0.6577 | Yes |
| 35 | TCF19 | TCF19 Entrez, Source | transcription factor 19 (SC1) | 12623 | -0.188 | -0.6543 | Yes |
| 36 | BIRC5 | BIRC5 Entrez, Source | baculoviral IAP repeat-containing 5 (survivin) | 12656 | -0.196 | -0.6370 | Yes |
| 37 | CDCA8 | CDCA8 Entrez, Source | cell division cycle associated 8 | 12816 | -0.228 | -0.6259 | Yes |
| 38 | ETS1 | ETS1 Entrez, Source | v-ets erythroblastosis virus E26 oncogene homolog 1 (avian) | 12930 | -0.262 | -0.6079 | Yes |
| 39 | CCNB2 | CCNB2 Entrez, Source | cyclin B2 | 12949 | -0.272 | -0.5818 | Yes |
| 40 | KIF22 | KIF22 Entrez, Source | kinesin family member 22 | 12985 | -0.285 | -0.5556 | Yes |
| 41 | HN1 | HN1 Entrez, Source | hematological and neurological expressed 1 | 13029 | -0.307 | -0.5279 | Yes |
| 42 | CCND1 | CCND1 Entrez, Source | cyclin D1 | 13085 | -0.343 | -0.4974 | Yes |
| 43 | LMNB1 | LMNB1 Entrez, Source | lamin B1 | 13086 | -0.343 | -0.4627 | Yes |
| 44 | BASP1 | BASP1 Entrez, Source | brain abundant, membrane attached signal protein 1 | 13112 | -0.366 | -0.4277 | Yes |
| 45 | CD44 | CD44 Entrez, Source | CD44 molecule (Indian blood group) | 13174 | -0.431 | -0.3887 | Yes |
| 46 | VIL2 | VIL2 Entrez, Source | villin 2 (ezrin) | 13200 | -0.467 | -0.3433 | Yes |
| 47 | CDC2A | 13232 | -0.520 | -0.2932 | Yes | ||
| 48 | CCNA2 | CCNA2 Entrez, Source | cyclin A2 | 13314 | -0.890 | -0.2094 | Yes |
| 49 | CCL2 | CCL2 Entrez, Source | chemokine (C-C motif) ligand 2 | 13317 | -0.925 | -0.1161 | Yes |
| 50 | MARCKSL1 | MARCKSL1 Entrez, Source | MARCKS-like 1 | 13330 | -1.167 | 0.0009 | Yes |