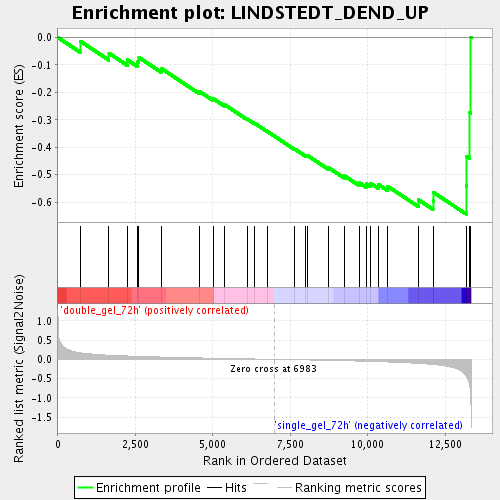
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_72h_versus_single_gel_72h.class.cls #double_gel_72h_versus_single_gel_72h_repos |
| Phenotype | class.cls#double_gel_72h_versus_single_gel_72h_repos |
| Upregulated in class | single_gel_72h |
| GeneSet | LINDSTEDT_DEND_UP |
| Enrichment Score (ES) | -0.64382637 |
| Normalized Enrichment Score (NES) | -1.7851743 |
| Nominal p-value | 0.0055762082 |
| FDR q-value | 0.039293237 |
| FWER p-Value | 0.987 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | CFLAR | CFLAR Entrez, Source | CASP8 and FADD-like apoptosis regulator | 717 | 0.171 | -0.0131 | No |
| 2 | CD80 | CD80 Entrez, Source | CD80 molecule | 1647 | 0.107 | -0.0574 | No |
| 3 | NFKBIA | NFKBIA Entrez, Source | nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha | 2241 | 0.087 | -0.0813 | No |
| 4 | OPTN | OPTN Entrez, Source | optineurin | 2577 | 0.077 | -0.0881 | No |
| 5 | PLA2G6 | PLA2G6 Entrez, Source | phospholipase A2, group VI (cytosolic, calcium-independent) | 2615 | 0.076 | -0.0728 | No |
| 6 | SDC4 | SDC4 Entrez, Source | syndecan 4 (amphiglycan, ryudocan) | 3350 | 0.059 | -0.1139 | No |
| 7 | DUSP4 | DUSP4 Entrez, Source | dual specificity phosphatase 4 | 4576 | 0.037 | -0.1972 | No |
| 8 | NFKB1 | NFKB1 Entrez, Source | nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 (p105) | 5011 | 0.029 | -0.2229 | No |
| 9 | BTG1 | BTG1 Entrez, Source | B-cell translocation gene 1, anti-proliferative | 5391 | 0.023 | -0.2458 | No |
| 10 | LYN | LYN Entrez, Source | v-yes-1 Yamaguchi sarcoma viral related oncogene homolog | 6114 | 0.013 | -0.2970 | No |
| 11 | SIAH2 | SIAH2 Entrez, Source | seven in absentia homolog 2 (Drosophila) | 6351 | 0.009 | -0.3127 | No |
| 12 | ID2 | ID2 Entrez, Source | inhibitor of DNA binding 2, dominant negative helix-loop-helix protein | 6773 | 0.003 | -0.3436 | No |
| 13 | NR4A3 | NR4A3 Entrez, Source | nuclear receptor subfamily 4, group A, member 3 | 7647 | -0.010 | -0.4069 | No |
| 14 | PSME2 | PSME2 Entrez, Source | proteasome (prosome, macropain) activator subunit 2 (PA28 beta) | 7999 | -0.015 | -0.4296 | No |
| 15 | NFKB2 | NFKB2 Entrez, Source | nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) | 8044 | -0.015 | -0.4293 | No |
| 16 | NBN | NBN Entrez, Source | nibrin | 8726 | -0.027 | -0.4740 | No |
| 17 | IL2RA | IL2RA Entrez, Source | interleukin 2 receptor, alpha | 9245 | -0.037 | -0.5042 | No |
| 18 | CXCR4 | CXCR4 Entrez, Source | chemokine (C-X-C motif) receptor 4 | 9737 | -0.046 | -0.5300 | No |
| 19 | GADD45A | GADD45A Entrez, Source | growth arrest and DNA-damage-inducible, alpha | 9946 | -0.051 | -0.5335 | No |
| 20 | MARCKS | MARCKS Entrez, Source | myristoylated alanine-rich protein kinase C substrate | 10092 | -0.054 | -0.5315 | No |
| 21 | PNRC1 | PNRC1 Entrez, Source | proline-rich nuclear receptor coactivator 1 | 10356 | -0.060 | -0.5369 | No |
| 22 | LAMP3 | LAMP3 Entrez, Source | lysosomal-associated membrane protein 3 | 10638 | -0.068 | -0.5418 | No |
| 23 | BIRC3 | BIRC3 Entrez, Source | baculoviral IAP repeat-containing 3 | 11637 | -0.106 | -0.5915 | No |
| 24 | KLF5 | KLF5 Entrez, Source | Kruppel-like factor 5 (intestinal) | 12107 | -0.132 | -0.5954 | Yes |
| 25 | TAP1 | TAP1 Entrez, Source | transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) | 12128 | -0.133 | -0.5652 | Yes |
| 26 | PDGFA | PDGFA Entrez, Source | platelet-derived growth factor alpha polypeptide | 13176 | -0.433 | -0.5407 | Yes |
| 27 | LAMB3 | LAMB3 Entrez, Source | laminin, beta 3 | 13195 | -0.458 | -0.4329 | Yes |
| 28 | ADM | ADM Entrez, Source | adrenomedullin | 13287 | -0.697 | -0.2738 | Yes |
| 29 | MARCKSL1 | MARCKSL1 Entrez, Source | MARCKS-like 1 | 13330 | -1.167 | 0.0009 | Yes |

