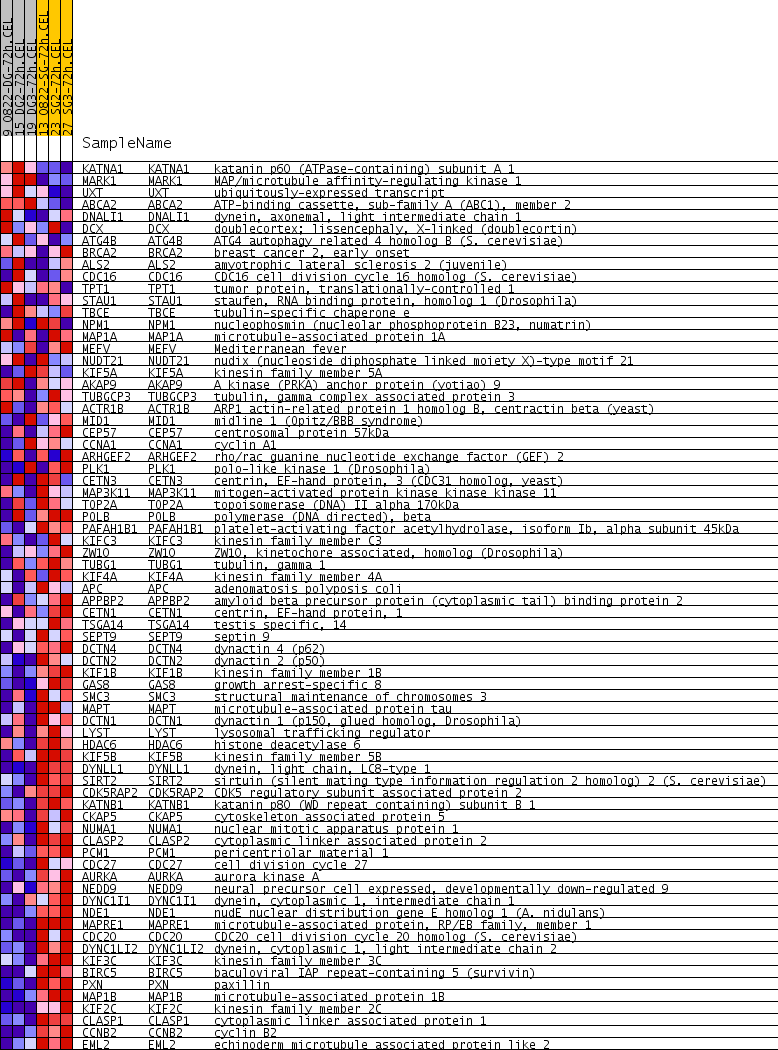
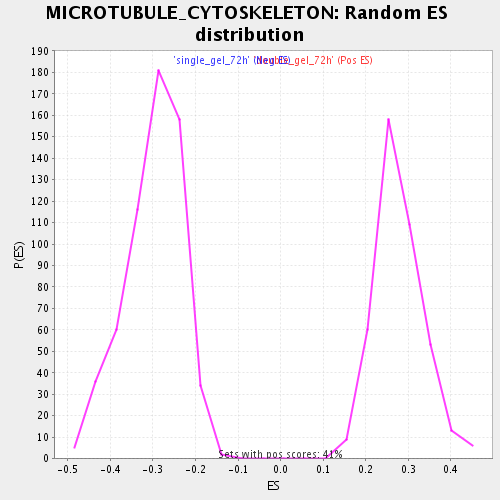
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_72h_versus_single_gel_72h.class.cls #double_gel_72h_versus_single_gel_72h_repos |
| Phenotype | class.cls#double_gel_72h_versus_single_gel_72h_repos |
| Upregulated in class | single_gel_72h |
| GeneSet | MICROTUBULE_CYTOSKELETON |
| Enrichment Score (ES) | -0.5793382 |
| Normalized Enrichment Score (NES) | -1.9441102 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.013190081 |
| FWER p-Value | 0.358 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | KATNA1 | KATNA1 Entrez, Source | katanin p60 (ATPase-containing) subunit A 1 | 1345 | 0.122 | -0.0807 | No |
| 2 | MARK1 | MARK1 Entrez, Source | MAP/microtubule affinity-regulating kinase 1 | 1653 | 0.107 | -0.0857 | No |
| 3 | UXT | UXT Entrez, Source | ubiquitously-expressed transcript | 3680 | 0.052 | -0.2295 | No |
| 4 | ABCA2 | ABCA2 Entrez, Source | ATP-binding cassette, sub-family A (ABC1), member 2 | 4097 | 0.045 | -0.2533 | No |
| 5 | DNALI1 | DNALI1 Entrez, Source | dynein, axonemal, light intermediate chain 1 | 4604 | 0.036 | -0.2853 | No |
| 6 | DCX | DCX Entrez, Source | doublecortex; lissencephaly, X-linked (doublecortin) | 4710 | 0.034 | -0.2874 | No |
| 7 | ATG4B | ATG4B Entrez, Source | ATG4 autophagy related 4 homolog B (S. cerevisiae) | 5885 | 0.016 | -0.3732 | No |
| 8 | BRCA2 | BRCA2 Entrez, Source | breast cancer 2, early onset | 5924 | 0.015 | -0.3734 | No |
| 9 | ALS2 | ALS2 Entrez, Source | amyotrophic lateral sclerosis 2 (juvenile) | 5959 | 0.015 | -0.3735 | No |
| 10 | CDC16 | CDC16 Entrez, Source | CDC16 cell division cycle 16 homolog (S. cerevisiae) | 6164 | 0.012 | -0.3868 | No |
| 11 | TPT1 | TPT1 Entrez, Source | tumor protein, translationally-controlled 1 | 6352 | 0.009 | -0.3994 | No |
| 12 | STAU1 | STAU1 Entrez, Source | staufen, RNA binding protein, homolog 1 (Drosophila) | 6669 | 0.004 | -0.4225 | No |
| 13 | TBCE | TBCE Entrez, Source | tubulin-specific chaperone e | 6963 | 0.000 | -0.4445 | No |
| 14 | NPM1 | NPM1 Entrez, Source | nucleophosmin (nucleolar phosphoprotein B23, numatrin) | 7074 | -0.001 | -0.4526 | No |
| 15 | MAP1A | MAP1A Entrez, Source | microtubule-associated protein 1A | 7116 | -0.002 | -0.4554 | No |
| 16 | MEFV | MEFV Entrez, Source | Mediterranean fever | 7182 | -0.003 | -0.4598 | No |
| 17 | NUDT21 | NUDT21 Entrez, Source | nudix (nucleoside diphosphate linked moiety X)-type motif 21 | 7285 | -0.004 | -0.4667 | No |
| 18 | KIF5A | KIF5A Entrez, Source | kinesin family member 5A | 7406 | -0.006 | -0.4747 | No |
| 19 | AKAP9 | AKAP9 Entrez, Source | A kinase (PRKA) anchor protein (yotiao) 9 | 7704 | -0.010 | -0.4953 | No |
| 20 | TUBGCP3 | TUBGCP3 Entrez, Source | tubulin, gamma complex associated protein 3 | 7754 | -0.011 | -0.4971 | No |
| 21 | ACTR1B | ACTR1B Entrez, Source | ARP1 actin-related protein 1 homolog B, centractin beta (yeast) | 7815 | -0.012 | -0.4995 | No |
| 22 | MID1 | MID1 Entrez, Source | midline 1 (Opitz/BBB syndrome) | 7962 | -0.014 | -0.5081 | No |
| 23 | CEP57 | CEP57 Entrez, Source | centrosomal protein 57kDa | 8017 | -0.015 | -0.5095 | No |
| 24 | CCNA1 | CCNA1 Entrez, Source | cyclin A1 | 8384 | -0.021 | -0.5335 | No |
| 25 | ARHGEF2 | ARHGEF2 Entrez, Source | rho/rac guanine nucleotide exchange factor (GEF) 2 | 8581 | -0.024 | -0.5441 | No |
| 26 | PLK1 | PLK1 Entrez, Source | polo-like kinase 1 (Drosophila) | 8736 | -0.027 | -0.5511 | No |
| 27 | CETN3 | CETN3 Entrez, Source | centrin, EF-hand protein, 3 (CDC31 homolog, yeast) | 8746 | -0.027 | -0.5472 | No |
| 28 | MAP3K11 | MAP3K11 Entrez, Source | mitogen-activated protein kinase kinase kinase 11 | 9043 | -0.033 | -0.5639 | No |
| 29 | TOP2A | TOP2A Entrez, Source | topoisomerase (DNA) II alpha 170kDa | 9129 | -0.035 | -0.5644 | No |
| 30 | POLB | POLB Entrez, Source | polymerase (DNA directed), beta | 9135 | -0.035 | -0.5589 | No |
| 31 | PAFAH1B1 | PAFAH1B1 Entrez, Source | platelet-activating factor acetylhydrolase, isoform Ib, alpha subunit 45kDa | 9229 | -0.036 | -0.5598 | No |
| 32 | KIFC3 | KIFC3 Entrez, Source | kinesin family member C3 | 9267 | -0.037 | -0.5562 | No |
| 33 | ZW10 | ZW10 Entrez, Source | ZW10, kinetochore associated, homolog (Drosophila) | 9270 | -0.037 | -0.5501 | No |
| 34 | TUBG1 | TUBG1 Entrez, Source | tubulin, gamma 1 | 9358 | -0.039 | -0.5500 | No |
| 35 | KIF4A | KIF4A Entrez, Source | kinesin family member 4A | 9748 | -0.047 | -0.5714 | Yes |
| 36 | APC | APC Entrez, Source | adenomatosis polyposis coli | 9845 | -0.049 | -0.5704 | Yes |
| 37 | APPBP2 | APPBP2 Entrez, Source | amyloid beta precursor protein (cytoplasmic tail) binding protein 2 | 9900 | -0.050 | -0.5659 | Yes |
| 38 | CETN1 | CETN1 Entrez, Source | centrin, EF-hand protein, 1 | 10032 | -0.053 | -0.5668 | Yes |
| 39 | TSGA14 | TSGA14 Entrez, Source | testis specific, 14 | 10166 | -0.056 | -0.5674 | Yes |
| 40 | SEPT9 | SEPT9 Entrez, Source | septin 9 | 10261 | -0.058 | -0.5646 | Yes |
| 41 | DCTN4 | DCTN4 Entrez, Source | dynactin 4 (p62) | 10318 | -0.060 | -0.5587 | Yes |
| 42 | DCTN2 | DCTN2 Entrez, Source | dynactin 2 (p50) | 10348 | -0.060 | -0.5506 | Yes |
| 43 | KIF1B | KIF1B Entrez, Source | kinesin family member 1B | 10358 | -0.060 | -0.5410 | Yes |
| 44 | GAS8 | GAS8 Entrez, Source | growth arrest-specific 8 | 10447 | -0.063 | -0.5370 | Yes |
| 45 | SMC3 | SMC3 Entrez, Source | structural maintenance of chromosomes 3 | 10686 | -0.069 | -0.5431 | Yes |
| 46 | MAPT | MAPT Entrez, Source | microtubule-associated protein tau | 10830 | -0.073 | -0.5415 | Yes |
| 47 | DCTN1 | DCTN1 Entrez, Source | dynactin 1 (p150, glued homolog, Drosophila) | 10978 | -0.078 | -0.5393 | Yes |
| 48 | LYST | LYST Entrez, Source | lysosomal trafficking regulator | 11059 | -0.080 | -0.5316 | Yes |
| 49 | HDAC6 | HDAC6 Entrez, Source | histone deacetylase 6 | 11173 | -0.085 | -0.5258 | Yes |
| 50 | KIF5B | KIF5B Entrez, Source | kinesin family member 5B | 11531 | -0.101 | -0.5356 | Yes |
| 51 | DYNLL1 | DYNLL1 Entrez, Source | dynein, light chain, LC8-type 1 | 11553 | -0.102 | -0.5198 | Yes |
| 52 | SIRT2 | SIRT2 Entrez, Source | sirtuin (silent mating type information regulation 2 homolog) 2 (S. cerevisiae) | 11576 | -0.103 | -0.5040 | Yes |
| 53 | CDK5RAP2 | CDK5RAP2 Entrez, Source | CDK5 regulatory subunit associated protein 2 | 11579 | -0.103 | -0.4866 | Yes |
| 54 | KATNB1 | KATNB1 Entrez, Source | katanin p80 (WD repeat containing) subunit B 1 | 11624 | -0.106 | -0.4720 | Yes |
| 55 | CKAP5 | CKAP5 Entrez, Source | cytoskeleton associated protein 5 | 11647 | -0.106 | -0.4555 | Yes |
| 56 | NUMA1 | NUMA1 Entrez, Source | nuclear mitotic apparatus protein 1 | 11712 | -0.110 | -0.4417 | Yes |
| 57 | CLASP2 | CLASP2 Entrez, Source | cytoplasmic linker associated protein 2 | 11749 | -0.111 | -0.4255 | Yes |
| 58 | PCM1 | PCM1 Entrez, Source | pericentriolar material 1 | 11900 | -0.119 | -0.4166 | Yes |
| 59 | CDC27 | CDC27 Entrez, Source | cell division cycle 27 | 11905 | -0.119 | -0.3968 | Yes |
| 60 | AURKA | AURKA Entrez, Source | aurora kinase A | 11917 | -0.119 | -0.3773 | Yes |
| 61 | NEDD9 | NEDD9 Entrez, Source | neural precursor cell expressed, developmentally down-regulated 9 | 11950 | -0.122 | -0.3591 | Yes |
| 62 | DYNC1I1 | DYNC1I1 Entrez, Source | dynein, cytoplasmic 1, intermediate chain 1 | 12058 | -0.129 | -0.3453 | Yes |
| 63 | NDE1 | NDE1 Entrez, Source | nudE nuclear distribution gene E homolog 1 (A. nidulans) | 12075 | -0.130 | -0.3245 | Yes |
| 64 | MAPRE1 | MAPRE1 Entrez, Source | microtubule-associated protein, RP/EB family, member 1 | 12109 | -0.132 | -0.3045 | Yes |
| 65 | CDC20 | CDC20 Entrez, Source | CDC20 cell division cycle 20 homolog (S. cerevisiae) | 12256 | -0.144 | -0.2910 | Yes |
| 66 | DYNC1LI2 | DYNC1LI2 Entrez, Source | dynein, cytoplasmic 1, light intermediate chain 2 | 12539 | -0.177 | -0.2823 | Yes |
| 67 | KIF3C | KIF3C Entrez, Source | kinesin family member 3C | 12599 | -0.184 | -0.2554 | Yes |
| 68 | BIRC5 | BIRC5 Entrez, Source | baculoviral IAP repeat-containing 5 (survivin) | 12656 | -0.196 | -0.2263 | Yes |
| 69 | PXN | PXN Entrez, Source | paxillin | 12740 | -0.211 | -0.1967 | Yes |
| 70 | MAP1B | MAP1B Entrez, Source | microtubule-associated protein 1B | 12813 | -0.228 | -0.1634 | Yes |
| 71 | KIF2C | KIF2C Entrez, Source | kinesin family member 2C | 12815 | -0.228 | -0.1246 | Yes |
| 72 | CLASP1 | CLASP1 Entrez, Source | cytoplasmic linker associated protein 1 | 12914 | -0.257 | -0.0883 | Yes |
| 73 | CCNB2 | CCNB2 Entrez, Source | cyclin B2 | 12949 | -0.272 | -0.0446 | Yes |
| 74 | EML2 | EML2 Entrez, Source | echinoderm microtubule associated protein like 2 | 13178 | -0.436 | 0.0124 | Yes |