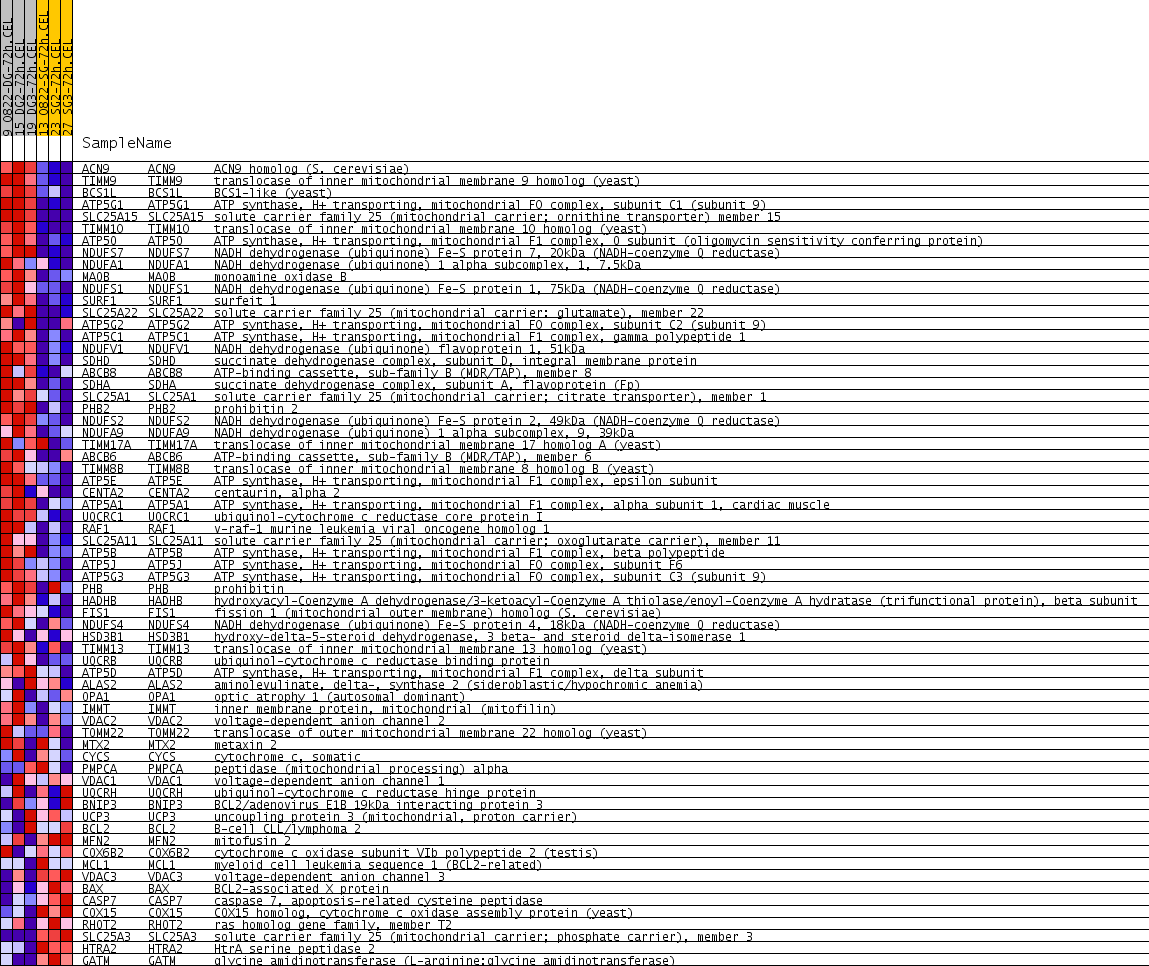
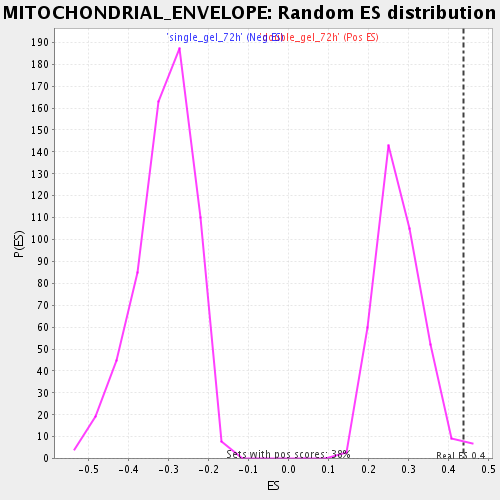
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_72h_versus_single_gel_72h.class.cls #double_gel_72h_versus_single_gel_72h_repos |
| Phenotype | class.cls#double_gel_72h_versus_single_gel_72h_repos |
| Upregulated in class | double_gel_72h |
| GeneSet | MITOCHONDRIAL_ENVELOPE |
| Enrichment Score (ES) | 0.4382222 |
| Normalized Enrichment Score (NES) | 1.581155 |
| Nominal p-value | 0.015831135 |
| FDR q-value | 0.0754969 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | ACN9 | ACN9 Entrez, Source | ACN9 homolog (S. cerevisiae) | 284 | 0.273 | 0.0413 | Yes |
| 2 | TIMM9 | TIMM9 Entrez, Source | translocase of inner mitochondrial membrane 9 homolog (yeast) | 463 | 0.212 | 0.0765 | Yes |
| 3 | BCS1L | BCS1L Entrez, Source | BCS1-like (yeast) | 504 | 0.201 | 0.1196 | Yes |
| 4 | ATP5G1 | ATP5G1 Entrez, Source | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C1 (subunit 9) | 561 | 0.191 | 0.1592 | Yes |
| 5 | SLC25A15 | SLC25A15 Entrez, Source | solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 15 | 595 | 0.186 | 0.1994 | Yes |
| 6 | TIMM10 | TIMM10 Entrez, Source | translocase of inner mitochondrial membrane 10 homolog (yeast) | 1021 | 0.143 | 0.2003 | Yes |
| 7 | ATP5O | ATP5O Entrez, Source | ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit (oligomycin sensitivity conferring protein) | 1360 | 0.121 | 0.2026 | Yes |
| 8 | NDUFS7 | NDUFS7 Entrez, Source | NADH dehydrogenase (ubiquinone) Fe-S protein 7, 20kDa (NADH-coenzyme Q reductase) | 1371 | 0.121 | 0.2295 | Yes |
| 9 | NDUFA1 | NDUFA1 Entrez, Source | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 1, 7.5kDa | 1663 | 0.106 | 0.2319 | Yes |
| 10 | MAOB | MAOB Entrez, Source | monoamine oxidase B | 1779 | 0.102 | 0.2466 | Yes |
| 11 | NDUFS1 | NDUFS1 Entrez, Source | NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) | 1817 | 0.101 | 0.2670 | Yes |
| 12 | SURF1 | SURF1 Entrez, Source | surfeit 1 | 1848 | 0.100 | 0.2876 | Yes |
| 13 | SLC25A22 | SLC25A22 Entrez, Source | solute carrier family 25 (mitochondrial carrier: glutamate), member 22 | 2140 | 0.090 | 0.2863 | Yes |
| 14 | ATP5G2 | ATP5G2 Entrez, Source | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C2 (subunit 9) | 2318 | 0.085 | 0.2923 | Yes |
| 15 | ATP5C1 | ATP5C1 Entrez, Source | ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 | 2437 | 0.082 | 0.3022 | Yes |
| 16 | NDUFV1 | NDUFV1 Entrez, Source | NADH dehydrogenase (ubiquinone) flavoprotein 1, 51kDa | 2468 | 0.081 | 0.3184 | Yes |
| 17 | SDHD | SDHD Entrez, Source | succinate dehydrogenase complex, subunit D, integral membrane protein | 2529 | 0.079 | 0.3320 | Yes |
| 18 | ABCB8 | ABCB8 Entrez, Source | ATP-binding cassette, sub-family B (MDR/TAP), member 8 | 2624 | 0.076 | 0.3422 | Yes |
| 19 | SDHA | SDHA Entrez, Source | succinate dehydrogenase complex, subunit A, flavoprotein (Fp) | 2780 | 0.072 | 0.3471 | Yes |
| 20 | SLC25A1 | SLC25A1 Entrez, Source | solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1 | 2912 | 0.069 | 0.3529 | Yes |
| 21 | PHB2 | PHB2 Entrez, Source | prohibitin 2 | 3135 | 0.064 | 0.3508 | Yes |
| 22 | NDUFS2 | NDUFS2 Entrez, Source | NADH dehydrogenase (ubiquinone) Fe-S protein 2, 49kDa (NADH-coenzyme Q reductase) | 3172 | 0.063 | 0.3625 | Yes |
| 23 | NDUFA9 | NDUFA9 Entrez, Source | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 9, 39kDa | 3253 | 0.061 | 0.3705 | Yes |
| 24 | TIMM17A | TIMM17A Entrez, Source | translocase of inner mitochondrial membrane 17 homolog A (yeast) | 3292 | 0.060 | 0.3815 | Yes |
| 25 | ABCB6 | ABCB6 Entrez, Source | ATP-binding cassette, sub-family B (MDR/TAP), member 6 | 3356 | 0.059 | 0.3902 | Yes |
| 26 | TIMM8B | TIMM8B Entrez, Source | translocase of inner mitochondrial membrane 8 homolog B (yeast) | 3559 | 0.055 | 0.3875 | Yes |
| 27 | ATP5E | ATP5E Entrez, Source | ATP synthase, H+ transporting, mitochondrial F1 complex, epsilon subunit | 3634 | 0.053 | 0.3942 | Yes |
| 28 | CENTA2 | CENTA2 Entrez, Source | centaurin, alpha 2 | 3650 | 0.053 | 0.4052 | Yes |
| 29 | ATP5A1 | ATP5A1 Entrez, Source | ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1, cardiac muscle | 3850 | 0.049 | 0.4014 | Yes |
| 30 | UQCRC1 | UQCRC1 Entrez, Source | ubiquinol-cytochrome c reductase core protein I | 3903 | 0.048 | 0.4086 | Yes |
| 31 | RAF1 | RAF1 Entrez, Source | v-raf-1 murine leukemia viral oncogene homolog 1 | 3941 | 0.048 | 0.4167 | Yes |
| 32 | SLC25A11 | SLC25A11 Entrez, Source | solute carrier family 25 (mitochondrial carrier; oxoglutarate carrier), member 11 | 4171 | 0.043 | 0.4094 | Yes |
| 33 | ATP5B | ATP5B Entrez, Source | ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide | 4193 | 0.043 | 0.4177 | Yes |
| 34 | ATP5J | ATP5J Entrez, Source | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit F6 | 4194 | 0.043 | 0.4276 | Yes |
| 35 | ATP5G3 | ATP5G3 Entrez, Source | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C3 (subunit 9) | 4222 | 0.043 | 0.4353 | Yes |
| 36 | PHB | PHB Entrez, Source | prohibitin | 4309 | 0.041 | 0.4382 | Yes |
| 37 | HADHB | HADHB Entrez, Source | hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (trifunctional protein), beta subunit | 4675 | 0.035 | 0.4187 | No |
| 38 | FIS1 | FIS1 Entrez, Source | fission 1 (mitochondrial outer membrane) homolog (S. cerevisiae) | 4704 | 0.034 | 0.4244 | No |
| 39 | NDUFS4 | NDUFS4 Entrez, Source | NADH dehydrogenase (ubiquinone) Fe-S protein 4, 18kDa (NADH-coenzyme Q reductase) | 4753 | 0.033 | 0.4285 | No |
| 40 | HSD3B1 | HSD3B1 Entrez, Source | hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 1 | 4997 | 0.029 | 0.4169 | No |
| 41 | TIMM13 | TIMM13 Entrez, Source | translocase of inner mitochondrial membrane 13 homolog (yeast) | 5146 | 0.027 | 0.4119 | No |
| 42 | UQCRB | UQCRB Entrez, Source | ubiquinol-cytochrome c reductase binding protein | 5231 | 0.026 | 0.4115 | No |
| 43 | ATP5D | ATP5D Entrez, Source | ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit | 5323 | 0.024 | 0.4102 | No |
| 44 | ALAS2 | ALAS2 Entrez, Source | aminolevulinate, delta-, synthase 2 (sideroblastic/hypochromic anemia) | 5606 | 0.020 | 0.3936 | No |
| 45 | OPA1 | OPA1 Entrez, Source | optic atrophy 1 (autosomal dominant) | 5620 | 0.020 | 0.3972 | No |
| 46 | IMMT | IMMT Entrez, Source | inner membrane protein, mitochondrial (mitofilin) | 5652 | 0.020 | 0.3994 | No |
| 47 | VDAC2 | VDAC2 Entrez, Source | voltage-dependent anion channel 2 | 5987 | 0.014 | 0.3775 | No |
| 48 | TOMM22 | TOMM22 Entrez, Source | translocase of outer mitochondrial membrane 22 homolog (yeast) | 6098 | 0.013 | 0.3722 | No |
| 49 | MTX2 | MTX2 Entrez, Source | metaxin 2 | 6206 | 0.011 | 0.3667 | No |
| 50 | CYCS | CYCS Entrez, Source | cytochrome c, somatic | 6707 | 0.004 | 0.3299 | No |
| 51 | PMPCA | PMPCA Entrez, Source | peptidase (mitochondrial processing) alpha | 7146 | -0.002 | 0.2974 | No |
| 52 | VDAC1 | VDAC1 Entrez, Source | voltage-dependent anion channel 1 | 7242 | -0.004 | 0.2911 | No |
| 53 | UQCRH | UQCRH Entrez, Source | ubiquinol-cytochrome c reductase hinge protein | 7358 | -0.005 | 0.2837 | No |
| 54 | BNIP3 | BNIP3 Entrez, Source | BCL2/adenovirus E1B 19kDa interacting protein 3 | 7875 | -0.013 | 0.2478 | No |
| 55 | UCP3 | UCP3 Entrez, Source | uncoupling protein 3 (mitochondrial, proton carrier) | 8164 | -0.017 | 0.2301 | No |
| 56 | BCL2 | BCL2 Entrez, Source | B-cell CLL/lymphoma 2 | 8887 | -0.030 | 0.1826 | No |
| 57 | MFN2 | MFN2 Entrez, Source | mitofusin 2 | 9004 | -0.032 | 0.1812 | No |
| 58 | COX6B2 | COX6B2 Entrez, Source | cytochrome c oxidase subunit VIb polypeptide 2 (testis) | 9208 | -0.036 | 0.1742 | No |
| 59 | MCL1 | MCL1 Entrez, Source | myeloid cell leukemia sequence 1 (BCL2-related) | 9244 | -0.037 | 0.1800 | No |
| 60 | VDAC3 | VDAC3 Entrez, Source | voltage-dependent anion channel 3 | 9603 | -0.043 | 0.1630 | No |
| 61 | BAX | BAX Entrez, Source | BCL2-associated X protein | 9773 | -0.047 | 0.1611 | No |
| 62 | CASP7 | CASP7 Entrez, Source | caspase 7, apoptosis-related cysteine peptidase | 10338 | -0.060 | 0.1324 | No |
| 63 | COX15 | COX15 Entrez, Source | COX15 homolog, cytochrome c oxidase assembly protein (yeast) | 10424 | -0.062 | 0.1402 | No |
| 64 | RHOT2 | RHOT2 Entrez, Source | ras homolog gene family, member T2 | 10650 | -0.068 | 0.1390 | No |
| 65 | SLC25A3 | SLC25A3 Entrez, Source | solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3 | 11129 | -0.083 | 0.1220 | No |
| 66 | HTRA2 | HTRA2 Entrez, Source | HtrA serine peptidase 2 | 11309 | -0.091 | 0.1294 | No |
| 67 | GATM | GATM Entrez, Source | glycine amidinotransferase (L-arginine:glycine amidinotransferase) | 11580 | -0.103 | 0.1327 | No |