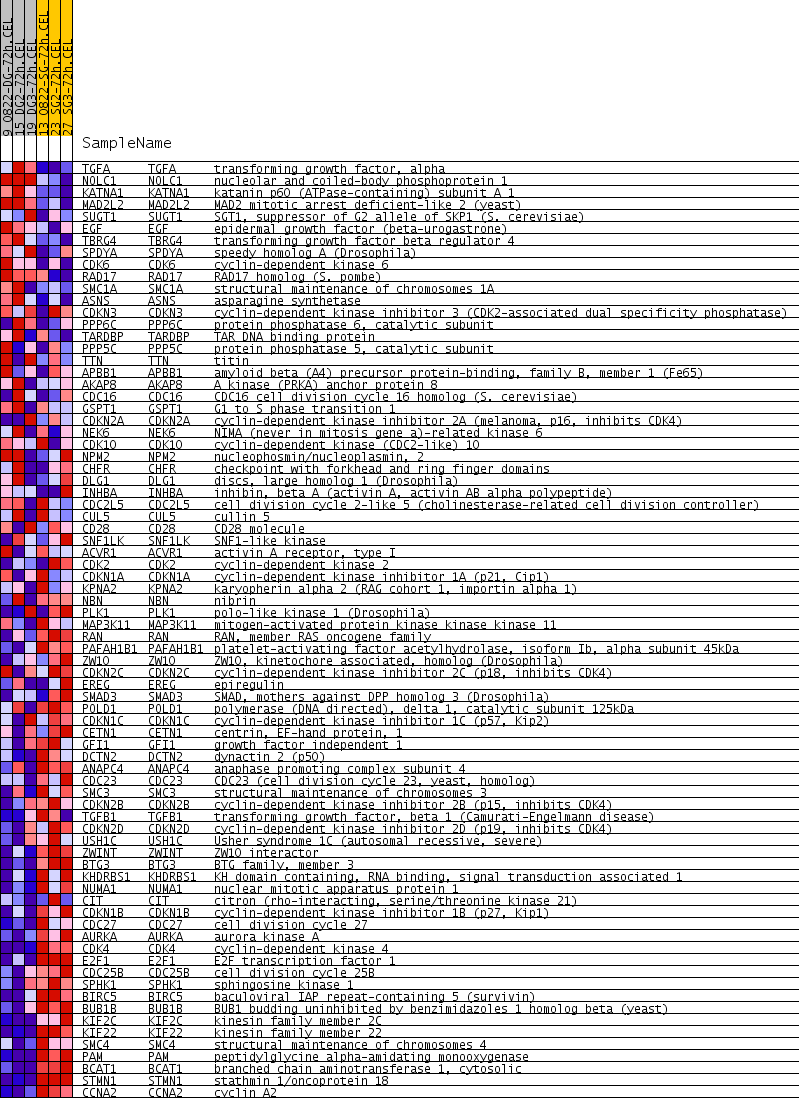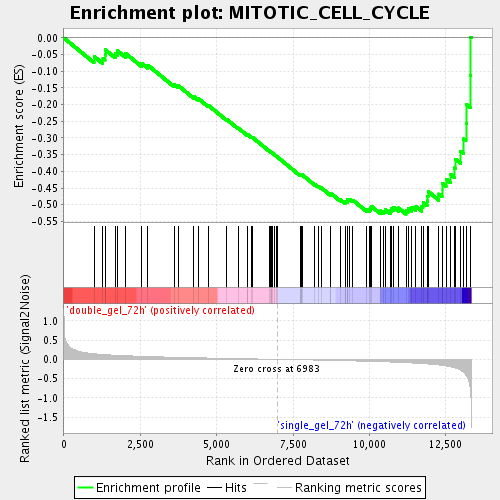
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_72h_versus_single_gel_72h.class.cls #double_gel_72h_versus_single_gel_72h_repos |
| Phenotype | class.cls#double_gel_72h_versus_single_gel_72h_repos |
| Upregulated in class | single_gel_72h |
| GeneSet | MITOTIC_CELL_CYCLE |
| Enrichment Score (ES) | -0.528192 |
| Normalized Enrichment Score (NES) | -1.7729506 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0423693 |
| FWER p-Value | 0.997 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | TGFA | TGFA Entrez, Source | transforming growth factor, alpha | 986 | 0.146 | -0.0555 | No |
| 2 | NOLC1 | NOLC1 Entrez, Source | nucleolar and coiled-body phosphoprotein 1 | 1277 | 0.125 | -0.0611 | No |
| 3 | KATNA1 | KATNA1 Entrez, Source | katanin p60 (ATPase-containing) subunit A 1 | 1345 | 0.122 | -0.0505 | No |
| 4 | MAD2L2 | MAD2L2 Entrez, Source | MAD2 mitotic arrest deficient-like 2 (yeast) | 1355 | 0.121 | -0.0356 | No |
| 5 | SUGT1 | SUGT1 Entrez, Source | SGT1, suppressor of G2 allele of SKP1 (S. cerevisiae) | 1681 | 0.106 | -0.0464 | No |
| 6 | EGF | EGF Entrez, Source | epidermal growth factor (beta-urogastrone) | 1748 | 0.103 | -0.0381 | No |
| 7 | TBRG4 | TBRG4 Entrez, Source | transforming growth factor beta regulator 4 | 2019 | 0.093 | -0.0464 | No |
| 8 | SPDYA | SPDYA Entrez, Source | speedy homolog A (Drosophila) | 2536 | 0.079 | -0.0752 | No |
| 9 | CDK6 | CDK6 Entrez, Source | cyclin-dependent kinase 6 | 2746 | 0.073 | -0.0816 | No |
| 10 | RAD17 | RAD17 Entrez, Source | RAD17 homolog (S. pombe) | 3607 | 0.054 | -0.1395 | No |
| 11 | SMC1A | SMC1A Entrez, Source | structural maintenance of chromosomes 1A | 3744 | 0.051 | -0.1432 | No |
| 12 | ASNS | ASNS Entrez, Source | asparagine synthetase | 4245 | 0.042 | -0.1754 | No |
| 13 | CDKN3 | CDKN3 Entrez, Source | cyclin-dependent kinase inhibitor 3 (CDK2-associated dual specificity phosphatase) | 4390 | 0.040 | -0.1812 | No |
| 14 | PPP6C | PPP6C Entrez, Source | protein phosphatase 6, catalytic subunit | 4715 | 0.034 | -0.2012 | No |
| 15 | TARDBP | TARDBP Entrez, Source | TAR DNA binding protein | 5332 | 0.024 | -0.2445 | No |
| 16 | PPP5C | PPP5C Entrez, Source | protein phosphatase 5, catalytic subunit | 5703 | 0.019 | -0.2700 | No |
| 17 | TTN | TTN Entrez, Source | titin | 5994 | 0.014 | -0.2900 | No |
| 18 | APBB1 | APBB1 Entrez, Source | amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) | 6000 | 0.014 | -0.2886 | No |
| 19 | AKAP8 | AKAP8 Entrez, Source | A kinase (PRKA) anchor protein 8 | 6151 | 0.012 | -0.2983 | No |
| 20 | CDC16 | CDC16 Entrez, Source | CDC16 cell division cycle 16 homolog (S. cerevisiae) | 6164 | 0.012 | -0.2977 | No |
| 21 | GSPT1 | GSPT1 Entrez, Source | G1 to S phase transition 1 | 6717 | 0.004 | -0.3389 | No |
| 22 | CDKN2A | CDKN2A Entrez, Source | cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4) | 6755 | 0.003 | -0.3413 | No |
| 23 | NEK6 | NEK6 Entrez, Source | NIMA (never in mitosis gene a)-related kinase 6 | 6778 | 0.003 | -0.3425 | No |
| 24 | CDK10 | CDK10 Entrez, Source | cyclin-dependent kinase (CDC2-like) 10 | 6796 | 0.003 | -0.3435 | No |
| 25 | NPM2 | NPM2 Entrez, Source | nucleophosmin/nucleoplasmin, 2 | 6828 | 0.002 | -0.3455 | No |
| 26 | CHFR | CHFR Entrez, Source | checkpoint with forkhead and ring finger domains | 6893 | 0.001 | -0.3502 | No |
| 27 | DLG1 | DLG1 Entrez, Source | discs, large homolog 1 (Drosophila) | 6958 | 0.000 | -0.3550 | No |
| 28 | INHBA | INHBA Entrez, Source | inhibin, beta A (activin A, activin AB alpha polypeptide) | 6984 | -0.000 | -0.3568 | No |
| 29 | CDC2L5 | CDC2L5 Entrez, Source | cell division cycle 2-like 5 (cholinesterase-related cell division controller) | 7727 | -0.011 | -0.4114 | No |
| 30 | CUL5 | CUL5 Entrez, Source | cullin 5 | 7748 | -0.011 | -0.4114 | No |
| 31 | CD28 | CD28 Entrez, Source | CD28 molecule | 7763 | -0.011 | -0.4110 | No |
| 32 | SNF1LK | SNF1LK Entrez, Source | SNF1-like kinase | 7785 | -0.012 | -0.4111 | No |
| 33 | ACVR1 | ACVR1 Entrez, Source | activin A receptor, type I | 7793 | -0.012 | -0.4101 | No |
| 34 | CDK2 | CDK2 Entrez, Source | cyclin-dependent kinase 2 | 8216 | -0.018 | -0.4395 | No |
| 35 | CDKN1A | CDKN1A Entrez, Source | cyclin-dependent kinase inhibitor 1A (p21, Cip1) | 8319 | -0.020 | -0.4446 | No |
| 36 | KPNA2 | KPNA2 Entrez, Source | karyopherin alpha 2 (RAG cohort 1, importin alpha 1) | 8416 | -0.022 | -0.4491 | No |
| 37 | NBN | NBN Entrez, Source | nibrin | 8726 | -0.027 | -0.4689 | No |
| 38 | PLK1 | PLK1 Entrez, Source | polo-like kinase 1 (Drosophila) | 8736 | -0.027 | -0.4661 | No |
| 39 | MAP3K11 | MAP3K11 Entrez, Source | mitogen-activated protein kinase kinase kinase 11 | 9043 | -0.033 | -0.4849 | No |
| 40 | RAN | RAN Entrez, Source | RAN, member RAS oncogene family | 9203 | -0.036 | -0.4923 | No |
| 41 | PAFAH1B1 | PAFAH1B1 Entrez, Source | platelet-activating factor acetylhydrolase, isoform Ib, alpha subunit 45kDa | 9229 | -0.036 | -0.4895 | No |
| 42 | ZW10 | ZW10 Entrez, Source | ZW10, kinetochore associated, homolog (Drosophila) | 9270 | -0.037 | -0.4877 | No |
| 43 | CDKN2C | CDKN2C Entrez, Source | cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4) | 9276 | -0.037 | -0.4832 | No |
| 44 | EREG | EREG Entrez, Source | epiregulin | 9343 | -0.039 | -0.4832 | No |
| 45 | SMAD3 | SMAD3 Entrez, Source | SMAD, mothers against DPP homolog 3 (Drosophila) | 9459 | -0.041 | -0.4866 | No |
| 46 | POLD1 | POLD1 Entrez, Source | polymerase (DNA directed), delta 1, catalytic subunit 125kDa | 9909 | -0.050 | -0.5140 | No |
| 47 | CDKN1C | CDKN1C Entrez, Source | cyclin-dependent kinase inhibitor 1C (p57, Kip2) | 10008 | -0.052 | -0.5147 | No |
| 48 | CETN1 | CETN1 Entrez, Source | centrin, EF-hand protein, 1 | 10032 | -0.053 | -0.5096 | No |
| 49 | GFI1 | GFI1 Entrez, Source | growth factor independent 1 | 10061 | -0.053 | -0.5048 | No |
| 50 | DCTN2 | DCTN2 Entrez, Source | dynactin 2 (p50) | 10348 | -0.060 | -0.5186 | No |
| 51 | ANAPC4 | ANAPC4 Entrez, Source | anaphase promoting complex subunit 4 | 10467 | -0.063 | -0.5193 | Yes |
| 52 | CDC23 | CDC23 Entrez, Source | CDC23 (cell division cycle 23, yeast, homolog) | 10523 | -0.065 | -0.5151 | Yes |
| 53 | SMC3 | SMC3 Entrez, Source | structural maintenance of chromosomes 3 | 10686 | -0.069 | -0.5184 | Yes |
| 54 | CDKN2B | CDKN2B Entrez, Source | cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4) | 10726 | -0.071 | -0.5122 | Yes |
| 55 | TGFB1 | TGFB1 Entrez, Source | transforming growth factor, beta 1 (Camurati-Engelmann disease) | 10788 | -0.072 | -0.5075 | Yes |
| 56 | CDKN2D | CDKN2D Entrez, Source | cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) | 10949 | -0.077 | -0.5096 | Yes |
| 57 | USH1C | USH1C Entrez, Source | Usher syndrome 1C (autosomal recessive, severe) | 11196 | -0.086 | -0.5172 | Yes |
| 58 | ZWINT | ZWINT Entrez, Source | ZW10 interactor | 11263 | -0.089 | -0.5107 | Yes |
| 59 | BTG3 | BTG3 Entrez, Source | BTG family, member 3 | 11377 | -0.094 | -0.5071 | Yes |
| 60 | KHDRBS1 | KHDRBS1 Entrez, Source | KH domain containing, RNA binding, signal transduction associated 1 | 11520 | -0.100 | -0.5049 | Yes |
| 61 | NUMA1 | NUMA1 Entrez, Source | nuclear mitotic apparatus protein 1 | 11712 | -0.110 | -0.5051 | Yes |
| 62 | CIT | CIT Entrez, Source | citron (rho-interacting, serine/threonine kinase 21) | 11752 | -0.111 | -0.4937 | Yes |
| 63 | CDKN1B | CDKN1B Entrez, Source | cyclin-dependent kinase inhibitor 1B (p27, Kip1) | 11886 | -0.118 | -0.4885 | Yes |
| 64 | CDC27 | CDC27 Entrez, Source | cell division cycle 27 | 11905 | -0.119 | -0.4745 | Yes |
| 65 | AURKA | AURKA Entrez, Source | aurora kinase A | 11917 | -0.119 | -0.4600 | Yes |
| 66 | CDK4 | CDK4 Entrez, Source | cyclin-dependent kinase 4 | 12272 | -0.146 | -0.4679 | Yes |
| 67 | E2F1 | E2F1 Entrez, Source | E2F transcription factor 1 | 12376 | -0.156 | -0.4556 | Yes |
| 68 | CDC25B | CDC25B Entrez, Source | cell division cycle 25B | 12377 | -0.156 | -0.4355 | Yes |
| 69 | SPHK1 | SPHK1 Entrez, Source | sphingosine kinase 1 | 12507 | -0.172 | -0.4230 | Yes |
| 70 | BIRC5 | BIRC5 Entrez, Source | baculoviral IAP repeat-containing 5 (survivin) | 12656 | -0.196 | -0.4089 | Yes |
| 71 | BUB1B | BUB1B Entrez, Source | BUB1 budding uninhibited by benzimidazoles 1 homolog beta (yeast) | 12774 | -0.218 | -0.3897 | Yes |
| 72 | KIF2C | KIF2C Entrez, Source | kinesin family member 2C | 12815 | -0.228 | -0.3632 | Yes |
| 73 | KIF22 | KIF22 Entrez, Source | kinesin family member 22 | 12985 | -0.285 | -0.3393 | Yes |
| 74 | SMC4 | SMC4 Entrez, Source | structural maintenance of chromosomes 4 | 13078 | -0.337 | -0.3027 | Yes |
| 75 | PAM | PAM Entrez, Source | peptidylglycine alpha-amidating monooxygenase | 13160 | -0.412 | -0.2556 | Yes |
| 76 | BCAT1 | BCAT1 Entrez, Source | branched chain aminotransferase 1, cytosolic | 13181 | -0.442 | -0.2002 | Yes |
| 77 | STMN1 | STMN1 Entrez, Source | stathmin 1/oncoprotein 18 | 13302 | -0.755 | -0.1118 | Yes |
| 78 | CCNA2 | CCNA2 Entrez, Source | cyclin A2 | 13314 | -0.890 | 0.0021 | Yes |