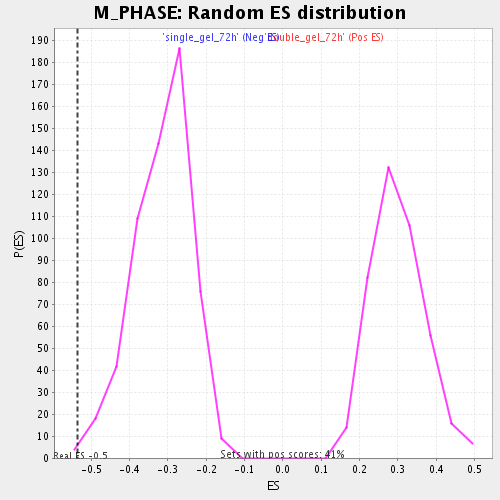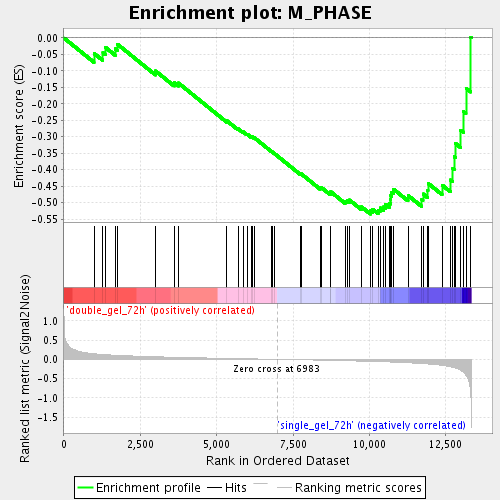
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_72h_versus_single_gel_72h.class.cls #double_gel_72h_versus_single_gel_72h_repos |
| Phenotype | class.cls#double_gel_72h_versus_single_gel_72h_repos |
| Upregulated in class | single_gel_72h |
| GeneSet | M_PHASE |
| Enrichment Score (ES) | -0.5349511 |
| Normalized Enrichment Score (NES) | -1.7003738 |
| Nominal p-value | 0.003407155 |
| FDR q-value | 0.06274015 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | TGFA | TGFA Entrez, Source | transforming growth factor, alpha | 986 | 0.146 | -0.0468 | No |
| 2 | NOLC1 | NOLC1 Entrez, Source | nucleolar and coiled-body phosphoprotein 1 | 1277 | 0.125 | -0.0452 | No |
| 3 | MAD2L2 | MAD2L2 Entrez, Source | MAD2 mitotic arrest deficient-like 2 (yeast) | 1355 | 0.121 | -0.0283 | No |
| 4 | SUGT1 | SUGT1 Entrez, Source | SGT1, suppressor of G2 allele of SKP1 (S. cerevisiae) | 1681 | 0.106 | -0.0330 | No |
| 5 | EGF | EGF Entrez, Source | epidermal growth factor (beta-urogastrone) | 1748 | 0.103 | -0.0187 | No |
| 6 | DUSP13 | DUSP13 Entrez, Source | dual specificity phosphatase 13 | 2997 | 0.067 | -0.1002 | No |
| 7 | RAD17 | RAD17 Entrez, Source | RAD17 homolog (S. pombe) | 3607 | 0.054 | -0.1360 | No |
| 8 | SMC1A | SMC1A Entrez, Source | structural maintenance of chromosomes 1A | 3744 | 0.051 | -0.1367 | No |
| 9 | TARDBP | TARDBP Entrez, Source | TAR DNA binding protein | 5332 | 0.024 | -0.2516 | No |
| 10 | PPP5C | PPP5C Entrez, Source | protein phosphatase 5, catalytic subunit | 5703 | 0.019 | -0.2759 | No |
| 11 | LIG3 | LIG3 Entrez, Source | ligase III, DNA, ATP-dependent | 5865 | 0.016 | -0.2850 | No |
| 12 | TTN | TTN Entrez, Source | titin | 5994 | 0.014 | -0.2920 | No |
| 13 | AKAP8 | AKAP8 Entrez, Source | A kinase (PRKA) anchor protein 8 | 6151 | 0.012 | -0.3014 | No |
| 14 | CDC16 | CDC16 Entrez, Source | CDC16 cell division cycle 16 homolog (S. cerevisiae) | 6164 | 0.012 | -0.3001 | No |
| 15 | MRE11A | MRE11A Entrez, Source | MRE11 meiotic recombination 11 homolog A (S. cerevisiae) | 6221 | 0.011 | -0.3023 | No |
| 16 | NEK6 | NEK6 Entrez, Source | NIMA (never in mitosis gene a)-related kinase 6 | 6778 | 0.003 | -0.3436 | No |
| 17 | NPM2 | NPM2 Entrez, Source | nucleophosmin/nucleoplasmin, 2 | 6828 | 0.002 | -0.3469 | No |
| 18 | CHFR | CHFR Entrez, Source | checkpoint with forkhead and ring finger domains | 6893 | 0.001 | -0.3514 | No |
| 19 | CDC2L5 | CDC2L5 Entrez, Source | cell division cycle 2-like 5 (cholinesterase-related cell division controller) | 7727 | -0.011 | -0.4121 | No |
| 20 | CD28 | CD28 Entrez, Source | CD28 molecule | 7763 | -0.011 | -0.4126 | No |
| 21 | CCNA1 | CCNA1 Entrez, Source | cyclin A1 | 8384 | -0.021 | -0.4552 | No |
| 22 | KPNA2 | KPNA2 Entrez, Source | karyopherin alpha 2 (RAG cohort 1, importin alpha 1) | 8416 | -0.022 | -0.4535 | No |
| 23 | NBN | NBN Entrez, Source | nibrin | 8726 | -0.027 | -0.4717 | No |
| 24 | PLK1 | PLK1 Entrez, Source | polo-like kinase 1 (Drosophila) | 8736 | -0.027 | -0.4674 | No |
| 25 | RAN | RAN Entrez, Source | RAN, member RAS oncogene family | 9203 | -0.036 | -0.4957 | No |
| 26 | ZW10 | ZW10 Entrez, Source | ZW10, kinetochore associated, homolog (Drosophila) | 9270 | -0.037 | -0.4937 | No |
| 27 | EREG | EREG Entrez, Source | epiregulin | 9343 | -0.039 | -0.4920 | No |
| 28 | SYCP1 | SYCP1 Entrez, Source | synaptonemal complex protein 1 | 9738 | -0.046 | -0.5129 | No |
| 29 | CETN1 | CETN1 Entrez, Source | centrin, EF-hand protein, 1 | 10032 | -0.053 | -0.5251 | Yes |
| 30 | HSPA2 | HSPA2 Entrez, Source | heat shock 70kDa protein 2 | 10104 | -0.054 | -0.5203 | Yes |
| 31 | SC65 | SC65 Entrez, Source | - | 10286 | -0.059 | -0.5229 | Yes |
| 32 | DCTN2 | DCTN2 Entrez, Source | dynactin 2 (p50) | 10348 | -0.060 | -0.5162 | Yes |
| 33 | ANAPC4 | ANAPC4 Entrez, Source | anaphase promoting complex subunit 4 | 10467 | -0.063 | -0.5132 | Yes |
| 34 | CDC23 | CDC23 Entrez, Source | CDC23 (cell division cycle 23, yeast, homolog) | 10523 | -0.065 | -0.5052 | Yes |
| 35 | STAG3 | STAG3 Entrez, Source | stromal antigen 3 | 10647 | -0.068 | -0.5017 | Yes |
| 36 | SMC3 | SMC3 Entrez, Source | structural maintenance of chromosomes 3 | 10686 | -0.069 | -0.4916 | Yes |
| 37 | RAD50 | RAD50 Entrez, Source | RAD50 homolog (S. cerevisiae) | 10695 | -0.070 | -0.4792 | Yes |
| 38 | CDKN2B | CDKN2B Entrez, Source | cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4) | 10726 | -0.071 | -0.4682 | Yes |
| 39 | TGFB1 | TGFB1 Entrez, Source | transforming growth factor, beta 1 (Camurati-Engelmann disease) | 10788 | -0.072 | -0.4593 | Yes |
| 40 | ZWINT | ZWINT Entrez, Source | ZW10 interactor | 11263 | -0.089 | -0.4784 | Yes |
| 41 | NUMA1 | NUMA1 Entrez, Source | nuclear mitotic apparatus protein 1 | 11712 | -0.110 | -0.4916 | Yes |
| 42 | CIT | CIT Entrez, Source | citron (rho-interacting, serine/threonine kinase 21) | 11752 | -0.111 | -0.4737 | Yes |
| 43 | CDC27 | CDC27 Entrez, Source | cell division cycle 27 | 11905 | -0.119 | -0.4629 | Yes |
| 44 | AURKA | AURKA Entrez, Source | aurora kinase A | 11917 | -0.119 | -0.4415 | Yes |
| 45 | CDC25B | CDC25B Entrez, Source | cell division cycle 25B | 12377 | -0.156 | -0.4469 | Yes |
| 46 | BIRC5 | BIRC5 Entrez, Source | baculoviral IAP repeat-containing 5 (survivin) | 12656 | -0.196 | -0.4312 | Yes |
| 47 | CHEK1 | CHEK1 Entrez, Source | CHK1 checkpoint homolog (S. pombe) | 12725 | -0.209 | -0.3972 | Yes |
| 48 | BUB1B | BUB1B Entrez, Source | BUB1 budding uninhibited by benzimidazoles 1 homolog beta (yeast) | 12774 | -0.218 | -0.3601 | Yes |
| 49 | KIF2C | KIF2C Entrez, Source | kinesin family member 2C | 12815 | -0.228 | -0.3204 | Yes |
| 50 | KIF22 | KIF22 Entrez, Source | kinesin family member 22 | 12985 | -0.285 | -0.2799 | Yes |
| 51 | SMC4 | SMC4 Entrez, Source | structural maintenance of chromosomes 4 | 13078 | -0.337 | -0.2238 | Yes |
| 52 | PAM | PAM Entrez, Source | peptidylglycine alpha-amidating monooxygenase | 13160 | -0.412 | -0.1528 | Yes |
| 53 | CCNA2 | CCNA2 Entrez, Source | cyclin A2 | 13314 | -0.890 | 0.0021 | Yes |