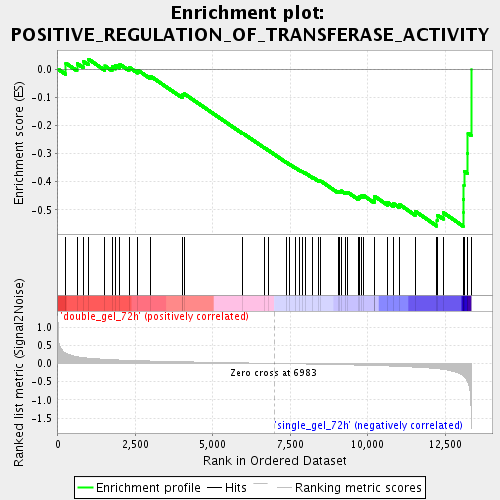
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_72h_versus_single_gel_72h.class.cls #double_gel_72h_versus_single_gel_72h_repos |
| Phenotype | class.cls#double_gel_72h_versus_single_gel_72h_repos |
| Upregulated in class | single_gel_72h |
| GeneSet | POSITIVE_REGULATION_OF_TRANSFERASE_ACTIVITY |
| Enrichment Score (ES) | -0.55806345 |
| Normalized Enrichment Score (NES) | -1.7404903 |
| Nominal p-value | 0.0016863407 |
| FDR q-value | 0.05021453 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | PRLR | PRLR Entrez, Source | prolactin receptor | 257 | 0.285 | 0.0208 | No |
| 2 | ADRB2 | ADRB2 Entrez, Source | adrenergic, beta-2-, receptor, surface | 617 | 0.184 | 0.0197 | No |
| 3 | GADD45G | GADD45G Entrez, Source | growth arrest and DNA-damage-inducible, gamma | 829 | 0.160 | 0.0263 | No |
| 4 | TGFA | TGFA Entrez, Source | transforming growth factor, alpha | 986 | 0.146 | 0.0351 | No |
| 5 | EDN2 | EDN2 Entrez, Source | endothelin 2 | 1518 | 0.112 | 0.0110 | No |
| 6 | EGF | EGF Entrez, Source | epidermal growth factor (beta-urogastrone) | 1748 | 0.103 | 0.0083 | No |
| 7 | GADD45B | GADD45B Entrez, Source | growth arrest and DNA-damage-inducible, beta | 1856 | 0.099 | 0.0142 | No |
| 8 | SHC1 | SHC1 Entrez, Source | SHC (Src homology 2 domain containing) transforming protein 1 | 1997 | 0.094 | 0.0169 | No |
| 9 | C5AR1 | C5AR1 Entrez, Source | complement component 5a receptor 1 | 2296 | 0.085 | 0.0064 | No |
| 10 | PICK1 | PICK1 Entrez, Source | protein interacting with PRKCA 1 | 2572 | 0.077 | -0.0034 | No |
| 11 | CD4 | CD4 Entrez, Source | CD4 molecule | 2984 | 0.067 | -0.0249 | No |
| 12 | CHRM1 | CHRM1 Entrez, Source | cholinergic receptor, muscarinic 1 | 4018 | 0.046 | -0.0961 | No |
| 13 | FGF2 | FGF2 Entrez, Source | fibroblast growth factor 2 (basic) | 4030 | 0.046 | -0.0905 | No |
| 14 | EGFR | EGFR Entrez, Source | epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | 4079 | 0.045 | -0.0878 | No |
| 15 | ALS2 | ALS2 Entrez, Source | amyotrophic lateral sclerosis 2 (juvenile) | 5959 | 0.015 | -0.2271 | No |
| 16 | MAP3K10 | MAP3K10 Entrez, Source | mitogen-activated protein kinase kinase kinase 10 | 6666 | 0.004 | -0.2796 | No |
| 17 | LYK5 | LYK5 Entrez, Source | - | 6784 | 0.003 | -0.2880 | No |
| 18 | CARTPT | CARTPT Entrez, Source | CART prepropeptide | 7367 | -0.006 | -0.3310 | No |
| 19 | GAP43 | GAP43 Entrez, Source | growth associated protein 43 | 7485 | -0.007 | -0.3388 | No |
| 20 | CHRNA7 | CHRNA7 Entrez, Source | cholinergic receptor, nicotinic, alpha 7 | 7665 | -0.010 | -0.3509 | No |
| 21 | SERINC1 | SERINC1 Entrez, Source | serine incorporator 1 | 7788 | -0.012 | -0.3584 | No |
| 22 | SOD1 | SOD1 Entrez, Source | superoxide dismutase 1, soluble (amyotrophic lateral sclerosis 1 (adult)) | 7883 | -0.013 | -0.3636 | No |
| 23 | GHRL | GHRL Entrez, Source | ghrelin/obestatin preprohormone | 7982 | -0.015 | -0.3689 | No |
| 24 | PPAP2A | PPAP2A Entrez, Source | phosphatidic acid phosphatase type 2A | 8226 | -0.018 | -0.3846 | No |
| 25 | PIK3CB | PIK3CB Entrez, Source | phosphoinositide-3-kinase, catalytic, beta polypeptide | 8414 | -0.022 | -0.3956 | No |
| 26 | DAXX | DAXX Entrez, Source | death-associated protein 6 | 8480 | -0.023 | -0.3973 | No |
| 27 | MAP3K11 | MAP3K11 Entrez, Source | mitogen-activated protein kinase kinase kinase 11 | 9043 | -0.033 | -0.4350 | No |
| 28 | GNG3 | GNG3 Entrez, Source | guanine nucleotide binding protein (G protein), gamma 3 | 9082 | -0.034 | -0.4331 | No |
| 29 | GRM4 | GRM4 Entrez, Source | glutamate receptor, metabotropic 4 | 9141 | -0.035 | -0.4326 | No |
| 30 | PAK1 | PAK1 Entrez, Source | p21/Cdc42/Rac1-activated kinase 1 (STE20 homolog, yeast) | 9287 | -0.038 | -0.4382 | No |
| 31 | EREG | EREG Entrez, Source | epiregulin | 9343 | -0.039 | -0.4369 | No |
| 32 | MADD | MADD Entrez, Source | MAP-kinase activating death domain | 9699 | -0.046 | -0.4572 | No |
| 33 | CXCR4 | CXCR4 Entrez, Source | chemokine (C-X-C motif) receptor 4 | 9737 | -0.046 | -0.4534 | No |
| 34 | MAP2K3 | MAP2K3 Entrez, Source | mitogen-activated protein kinase kinase 3 | 9785 | -0.048 | -0.4503 | No |
| 35 | CCND3 | CCND3 Entrez, Source | cyclin D3 | 9869 | -0.049 | -0.4496 | No |
| 36 | DBNL | DBNL Entrez, Source | drebrin-like | 10207 | -0.057 | -0.4670 | No |
| 37 | TAOK2 | TAOK2 Entrez, Source | TAO kinase 2 | 10217 | -0.057 | -0.4596 | No |
| 38 | ADRA2C | ADRA2C Entrez, Source | adrenergic, alpha-2C-, receptor | 10223 | -0.057 | -0.4519 | No |
| 39 | PARD3 | PARD3 Entrez, Source | par-3 partitioning defective 3 homolog (C. elegans) | 10642 | -0.068 | -0.4738 | No |
| 40 | ADRA2A | ADRA2A Entrez, Source | adrenergic, alpha-2A-, receptor | 10829 | -0.073 | -0.4775 | No |
| 41 | CD81 | CD81 Entrez, Source | CD81 molecule | 11022 | -0.079 | -0.4808 | No |
| 42 | PKN1 | PKN1 Entrez, Source | protein kinase N1 | 11530 | -0.101 | -0.5048 | No |
| 43 | ERBB2 | ERBB2 Entrez, Source | v-erb-b2 erythroblastic leukemia viral oncogene homolog 2, neuro/glioblastoma derived oncogene homolog (avian) | 12233 | -0.142 | -0.5376 | Yes |
| 44 | EDG5 | EDG5 Entrez, Source | endothelial differentiation, sphingolipid G-protein-coupled receptor, 5 | 12247 | -0.143 | -0.5184 | Yes |
| 45 | PROK2 | PROK2 Entrez, Source | prokineticin 2 | 12439 | -0.162 | -0.5099 | Yes |
| 46 | CCND2 | CCND2 Entrez, Source | cyclin D2 | 13080 | -0.340 | -0.5102 | Yes |
| 47 | CCND1 | CCND1 Entrez, Source | cyclin D1 | 13085 | -0.343 | -0.4623 | Yes |
| 48 | ADORA2B | ADORA2B Entrez, Source | adenosine A2b receptor | 13103 | -0.358 | -0.4132 | Yes |
| 49 | PLCE1 | PLCE1 Entrez, Source | phospholipase C, epsilon 1 | 13110 | -0.363 | -0.3625 | Yes |
| 50 | SERINC5 | SERINC5 Entrez, Source | serine incorporator 5 | 13225 | -0.503 | -0.3002 | Yes |
| 51 | SERINC2 | SERINC2 Entrez, Source | serine incorporator 2 | 13234 | -0.520 | -0.2276 | Yes |
| 52 | CD24 | CD24 Entrez, Source | CD24 molecule | 13341 | -1.675 | 0.0001 | Yes |