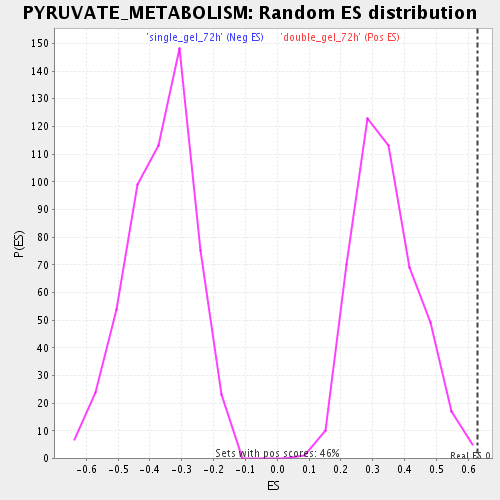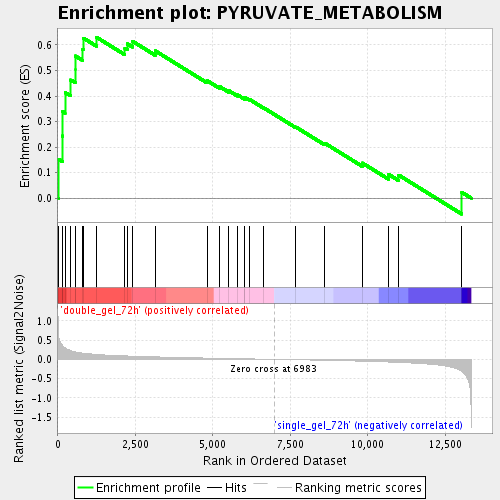
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_72h_versus_single_gel_72h.class.cls #double_gel_72h_versus_single_gel_72h_repos |
| Phenotype | class.cls#double_gel_72h_versus_single_gel_72h_repos |
| Upregulated in class | double_gel_72h |
| GeneSet | PYRUVATE_METABOLISM |
| Enrichment Score (ES) | 0.63030916 |
| Normalized Enrichment Score (NES) | 1.8441681 |
| Nominal p-value | 0.0021881838 |
| FDR q-value | 0.010752326 |
| FWER p-Value | 0.728 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | PKLR | PKLR Entrez, Source | pyruvate kinase, liver and RBC | 31 | 0.561 | 0.1524 | Yes |
| 2 | ACAT2 | ACAT2 Entrez, Source | acetyl-Coenzyme A acetyltransferase 2 (acetoacetyl Coenzyme A thiolase) | 144 | 0.362 | 0.2438 | Yes |
| 3 | PC | PC Entrez, Source | pyruvate carboxylase | 155 | 0.347 | 0.3388 | Yes |
| 4 | ACACA | ACACA Entrez, Source | acetyl-Coenzyme A carboxylase alpha | 231 | 0.293 | 0.4140 | Yes |
| 5 | PDHA2 | PDHA2 Entrez, Source | pyruvate dehydrogenase (lipoamide) alpha 2 | 414 | 0.227 | 0.4630 | Yes |
| 6 | ACAT1 | ACAT1 Entrez, Source | acetyl-Coenzyme A acetyltransferase 1 (acetoacetyl Coenzyme A thiolase) | 560 | 0.191 | 0.5047 | Yes |
| 7 | PDHB | PDHB Entrez, Source | pyruvate dehydrogenase (lipoamide) beta | 563 | 0.190 | 0.5571 | Yes |
| 8 | ME1 | ME1 Entrez, Source | malic enzyme 1, NADP(+)-dependent, cytosolic | 800 | 0.163 | 0.5843 | Yes |
| 9 | HAGH | HAGH Entrez, Source | hydroxyacylglutathione hydrolase | 819 | 0.161 | 0.6273 | Yes |
| 10 | MDH1 | MDH1 Entrez, Source | malate dehydrogenase 1, NAD (soluble) | 1246 | 0.127 | 0.6303 | Yes |
| 11 | LDHD | LDHD Entrez, Source | lactate dehydrogenase D | 2160 | 0.089 | 0.5864 | No |
| 12 | ALDH1A1 | ALDH1A1 Entrez, Source | aldehyde dehydrogenase 1 family, member A1 | 2239 | 0.087 | 0.6045 | No |
| 13 | MDH2 | MDH2 Entrez, Source | malate dehydrogenase 2, NAD (mitochondrial) | 2409 | 0.082 | 0.6145 | No |
| 14 | DLD | DLD Entrez, Source | dihydrolipoamide dehydrogenase | 3142 | 0.063 | 0.5771 | No |
| 15 | ALDH3A2 | ALDH3A2 Entrez, Source | aldehyde dehydrogenase 3 family, member A2 | 4819 | 0.032 | 0.4600 | No |
| 16 | ALDH1B1 | ALDH1B1 Entrez, Source | aldehyde dehydrogenase 1 family, member B1 | 5218 | 0.026 | 0.4373 | No |
| 17 | ALDH1A2 | ALDH1A2 Entrez, Source | aldehyde dehydrogenase 1 family, member A2 | 5520 | 0.021 | 0.4206 | No |
| 18 | PDHA1 | PDHA1 Entrez, Source | pyruvate dehydrogenase (lipoamide) alpha 1 | 5797 | 0.017 | 0.4047 | No |
| 19 | PCK1 | PCK1 Entrez, Source | phosphoenolpyruvate carboxykinase 1 (soluble) | 6026 | 0.014 | 0.3914 | No |
| 20 | GLO1 | GLO1 Entrez, Source | glyoxalase I | 6031 | 0.014 | 0.3949 | No |
| 21 | ALDH2 | ALDH2 Entrez, Source | aldehyde dehydrogenase 2 family (mitochondrial) | 6175 | 0.012 | 0.3874 | No |
| 22 | LDHC | LDHC Entrez, Source | lactate dehydrogenase C | 6640 | 0.005 | 0.3539 | No |
| 23 | LDHA | LDHA Entrez, Source | lactate dehydrogenase A | 7676 | -0.010 | 0.2789 | No |
| 24 | HAGHL | HAGHL Entrez, Source | hydroxyacylglutathione hydrolase-like | 8605 | -0.025 | 0.2161 | No |
| 25 | ALDH3A1 | ALDH3A1 Entrez, Source | aldehyde dehydrogenase 3 family, memberA1 | 9824 | -0.048 | 0.1379 | No |
| 26 | ALDH9A1 | ALDH9A1 Entrez, Source | aldehyde dehydrogenase 9 family, member A1 | 10674 | -0.069 | 0.0932 | No |
| 27 | DLAT | DLAT Entrez, Source | dihydrolipoamide S-acetyltransferase (E2 component of pyruvate dehydrogenase complex) | 10995 | -0.078 | 0.0908 | No |
| 28 | LDHB | LDHB Entrez, Source | lactate dehydrogenase B | 13034 | -0.309 | 0.0231 | No |