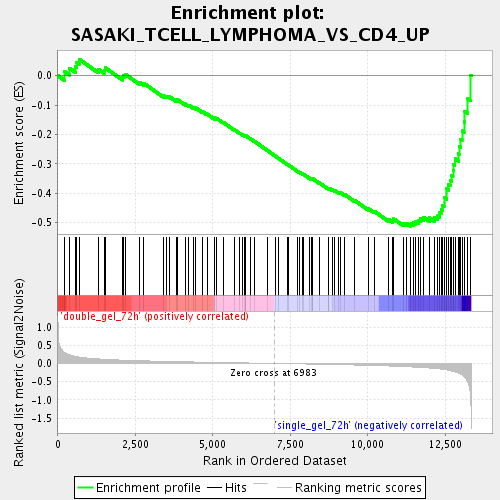
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_72h_versus_single_gel_72h.class.cls #double_gel_72h_versus_single_gel_72h_repos |
| Phenotype | class.cls#double_gel_72h_versus_single_gel_72h_repos |
| Upregulated in class | single_gel_72h |
| GeneSet | SASAKI_TCELL_LYMPHOMA_VS_CD4_UP |
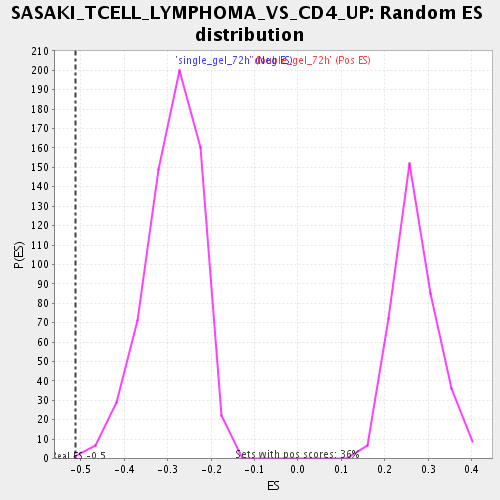
| Enrichment Score (ES) | -0.5122487 |
| Normalized Enrichment Score (NES) | -1.7722397 |
| Nominal p-value | 0.0015649452 |
| FDR q-value | 0.04140764 |
| FWER p-Value | 0.997 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | CEBPA | CEBPA Entrez, Source | CCAAT/enhancer binding protein (C/EBP), alpha | 203 | 0.308 | 0.0148 | No |
| 2 | HSD11B1 | HSD11B1 Entrez, Source | hydroxysteroid (11-beta) dehydrogenase 1 | 370 | 0.242 | 0.0259 | No |
| 3 | GCHFR | GCHFR Entrez, Source | GTP cyclohydrolase I feedback regulator | 558 | 0.191 | 0.0305 | No |
| 4 | TFRC | TFRC Entrez, Source | transferrin receptor (p90, CD71) | 608 | 0.185 | 0.0449 | No |
| 5 | DHCR24 | DHCR24 Entrez, Source | 24-dehydrocholesterol reductase | 685 | 0.175 | 0.0563 | No |
| 6 | ISG20 | ISG20 Entrez, Source | interferon stimulated exonuclease gene 20kDa | 1307 | 0.124 | 0.0215 | No |
| 7 | IDH1 | IDH1 Entrez, Source | isocitrate dehydrogenase 1 (NADP+), soluble | 1494 | 0.114 | 0.0186 | No |
| 8 | FAS | FAS Entrez, Source | Fas (TNF receptor superfamily, member 6) | 1520 | 0.112 | 0.0277 | No |
| 9 | KCNN4 | KCNN4 Entrez, Source | potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 | 2085 | 0.091 | -0.0060 | No |
| 10 | JAK3 | JAK3 Entrez, Source | Janus kinase 3 (a protein tyrosine kinase, leukocyte) | 2109 | 0.091 | 0.0012 | No |
| 11 | ACOT8 | ACOT8 Entrez, Source | acyl-CoA thioesterase 8 | 2184 | 0.089 | 0.0043 | No |
| 12 | PTPRM | PTPRM Entrez, Source | protein tyrosine phosphatase, receptor type, M | 2639 | 0.075 | -0.0227 | No |
| 13 | NME1 | NME1 Entrez, Source | non-metastatic cells 1, protein (NM23A) expressed in | 2764 | 0.072 | -0.0249 | No |
| 14 | STAT1 | STAT1 Entrez, Source | signal transducer and activator of transcription 1, 91kDa | 3416 | 0.058 | -0.0684 | No |
| 15 | CSNK1G1 | CSNK1G1 Entrez, Source | casein kinase 1, gamma 1 | 3501 | 0.056 | -0.0693 | No |
| 16 | TMED3 | TMED3 Entrez, Source | transmembrane emp24 protein transport domain containing 3 | 3590 | 0.054 | -0.0707 | No |
| 17 | SCCPDH | SCCPDH Entrez, Source | saccharopine dehydrogenase (putative) | 3830 | 0.049 | -0.0839 | No |
| 18 | ATP5A1 | ATP5A1 Entrez, Source | ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1, cardiac muscle | 3850 | 0.049 | -0.0805 | No |
| 19 | POU4F1 | POU4F1 Entrez, Source | POU domain, class 4, transcription factor 1 | 4112 | 0.044 | -0.0959 | No |
| 20 | ATP5G3 | ATP5G3 Entrez, Source | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C3 (subunit 9) | 4222 | 0.043 | -0.1000 | No |
| 21 | CDKN3 | CDKN3 Entrez, Source | cyclin-dependent kinase inhibitor 3 (CDK2-associated dual specificity phosphatase) | 4390 | 0.040 | -0.1087 | No |
| 22 | PPIB | PPIB Entrez, Source | peptidylprolyl isomerase B (cyclophilin B) | 4455 | 0.039 | -0.1097 | No |
| 23 | WDR18 | WDR18 Entrez, Source | WD repeat domain 18 | 4678 | 0.035 | -0.1231 | No |
| 24 | PTPRN2 | PTPRN2 Entrez, Source | protein tyrosine phosphatase, receptor type, N polypeptide 2 | 4815 | 0.032 | -0.1302 | No |
| 25 | MSX1 | MSX1 Entrez, Source | msh homeobox homolog 1 (Drosophila) | 5059 | 0.028 | -0.1458 | No |
| 26 | UBA52 | UBA52 Entrez, Source | ubiquitin A-52 residue ribosomal protein fusion product 1 | 5068 | 0.028 | -0.1437 | No |
| 27 | PTHLH | PTHLH Entrez, Source | parathyroid hormone-like hormone | 5115 | 0.027 | -0.1445 | No |
| 28 | ETFA | ETFA Entrez, Source | electron-transfer-flavoprotein, alpha polypeptide (glutaric aciduria II) | 5336 | 0.024 | -0.1587 | No |
| 29 | THRAP4 | THRAP4 Entrez, Source | thyroid hormone receptor associated protein 4 | 5700 | 0.019 | -0.1843 | No |
| 30 | ARNT2 | ARNT2 Entrez, Source | aryl-hydrocarbon receptor nuclear translocator 2 | 5871 | 0.016 | -0.1955 | No |
| 31 | PCNA | PCNA Entrez, Source | proliferating cell nuclear antigen | 5968 | 0.015 | -0.2013 | No |
| 32 | ICMT | ICMT Entrez, Source | isoprenylcysteine carboxyl methyltransferase | 6035 | 0.014 | -0.2050 | No |
| 33 | FGFR1 | FGFR1 Entrez, Source | fibroblast growth factor receptor 1 (fms-related tyrosine kinase 2, Pfeiffer syndrome) | 6041 | 0.014 | -0.2040 | No |
| 34 | PSMB2 | PSMB2 Entrez, Source | proteasome (prosome, macropain) subunit, beta type, 2 | 6045 | 0.014 | -0.2029 | No |
| 35 | MTX2 | MTX2 Entrez, Source | metaxin 2 | 6206 | 0.011 | -0.2139 | No |
| 36 | NMI | NMI Entrez, Source | N-myc (and STAT) interactor | 6331 | 0.009 | -0.2224 | No |
| 37 | HINT1 | HINT1 Entrez, Source | histidine triad nucleotide binding protein 1 | 6763 | 0.003 | -0.2546 | No |
| 38 | RECQL | RECQL Entrez, Source | RecQ protein-like (DNA helicase Q1-like) | 7030 | -0.001 | -0.2746 | No |
| 39 | PPID | PPID Entrez, Source | peptidylprolyl isomerase D (cyclophilin D) | 7131 | -0.002 | -0.2820 | No |
| 40 | ATP1B1 | ATP1B1 Entrez, Source | ATPase, Na+/K+ transporting, beta 1 polypeptide | 7404 | -0.006 | -0.3019 | No |
| 41 | SMARCA2 | SMARCA2 Entrez, Source | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 | 7445 | -0.007 | -0.3043 | No |
| 42 | CDC2L5 | CDC2L5 Entrez, Source | cell division cycle 2-like 5 (cholinesterase-related cell division controller) | 7727 | -0.011 | -0.3244 | No |
| 43 | PLD1 | PLD1 Entrez, Source | phospholipase D1, phosphatidylcholine-specific | 7809 | -0.012 | -0.3293 | No |
| 44 | SSTR2 | SSTR2 Entrez, Source | somatostatin receptor 2 | 7882 | -0.013 | -0.3335 | No |
| 45 | EXTL2 | EXTL2 Entrez, Source | exostoses (multiple)-like 2 | 7923 | -0.014 | -0.3351 | No |
| 46 | SP1 | SP1 Entrez, Source | Sp1 transcription factor | 8115 | -0.016 | -0.3479 | No |
| 47 | PDE8B | PDE8B Entrez, Source | phosphodiesterase 8B | 8180 | -0.018 | -0.3510 | No |
| 48 | PPP1CC | PPP1CC Entrez, Source | protein phosphatase 1, catalytic subunit, gamma isoform | 8221 | -0.018 | -0.3523 | No |
| 49 | PARP1 | PARP1 Entrez, Source | poly (ADP-ribose) polymerase family, member 1 | 8224 | -0.018 | -0.3506 | No |
| 50 | CORO1A | CORO1A Entrez, Source | coronin, actin binding protein, 1A | 8440 | -0.022 | -0.3647 | No |
| 51 | PLK1 | PLK1 Entrez, Source | polo-like kinase 1 (Drosophila) | 8736 | -0.027 | -0.3843 | No |
| 52 | CETN3 | CETN3 Entrez, Source | centrin, EF-hand protein, 3 (CDC31 homolog, yeast) | 8746 | -0.027 | -0.3823 | No |
| 53 | SELL | SELL Entrez, Source | selectin L (lymphocyte adhesion molecule 1) | 8867 | -0.029 | -0.3885 | No |
| 54 | SYT11 | SYT11 Entrez, Source | synaptotagmin XI | 8937 | -0.031 | -0.3907 | No |
| 55 | ANXA6 | ANXA6 Entrez, Source | annexin A6 | 9062 | -0.033 | -0.3968 | No |
| 56 | TOP2A | TOP2A Entrez, Source | topoisomerase (DNA) II alpha 170kDa | 9129 | -0.035 | -0.3984 | No |
| 57 | IL2RA | IL2RA Entrez, Source | interleukin 2 receptor, alpha | 9245 | -0.037 | -0.4035 | No |
| 58 | PRDX1 | PRDX1 Entrez, Source | peroxiredoxin 1 | 9564 | -0.043 | -0.4233 | No |
| 59 | PRDX4 | PRDX4 Entrez, Source | peroxiredoxin 4 | 10020 | -0.053 | -0.4525 | No |
| 60 | PPP1CA | PPP1CA Entrez, Source | protein phosphatase 1, catalytic subunit, alpha isoform | 10210 | -0.057 | -0.4612 | No |
| 61 | TRIB1 | TRIB1 Entrez, Source | tribbles homolog 1 (Drosophila) | 10667 | -0.069 | -0.4889 | No |
| 62 | FOXM1 | FOXM1 Entrez, Source | forkhead box M1 | 10798 | -0.072 | -0.4916 | No |
| 63 | MYO1D | MYO1D Entrez, Source | myosin ID | 10838 | -0.073 | -0.4874 | No |
| 64 | BARD1 | BARD1 Entrez, Source | BRCA1 associated RING domain 1 | 11154 | -0.084 | -0.5030 | No |
| 65 | ZWINT | ZWINT Entrez, Source | ZW10 interactor | 11263 | -0.089 | -0.5024 | No |
| 66 | VAMP5 | VAMP5 Entrez, Source | vesicle-associated membrane protein 5 (myobrevin) | 11394 | -0.095 | -0.5030 | Yes |
| 67 | CD3D | CD3D Entrez, Source | CD3d molecule, delta (CD3-TCR complex) | 11471 | -0.098 | -0.4991 | Yes |
| 68 | DYNLL1 | DYNLL1 Entrez, Source | dynein, light chain, LC8-type 1 | 11553 | -0.102 | -0.4952 | Yes |
| 69 | IGSF4 | IGSF4 Entrez, Source | immunoglobulin superfamily, member 4 | 11649 | -0.107 | -0.4920 | Yes |
| 70 | ING1 | ING1 Entrez, Source | inhibitor of growth family, member 1 | 11699 | -0.109 | -0.4850 | Yes |
| 71 | ATP2C1 | ATP2C1 Entrez, Source | ATPase, Ca++ transporting, type 2C, member 1 | 11799 | -0.114 | -0.4814 | Yes |
| 72 | RASA1 | RASA1 Entrez, Source | RAS p21 protein activator (GTPase activating protein) 1 | 11988 | -0.124 | -0.4834 | Yes |
| 73 | TRAFD1 | TRAFD1 Entrez, Source | TRAF-type zinc finger domain containing 1 | 12149 | -0.134 | -0.4824 | Yes |
| 74 | PAWR | PAWR Entrez, Source | PRKC, apoptosis, WT1, regulator | 12264 | -0.145 | -0.4768 | Yes |
| 75 | CD99 | CD99 Entrez, Source | CD99 molecule | 12323 | -0.150 | -0.4665 | Yes |
| 76 | SGCE | SGCE Entrez, Source | sarcoglycan, epsilon | 12368 | -0.155 | -0.4547 | Yes |
| 77 | FZD6 | FZD6 Entrez, Source | frizzled homolog 6 (Drosophila) | 12417 | -0.160 | -0.4426 | Yes |
| 78 | PTPRJ | PTPRJ Entrez, Source | protein tyrosine phosphatase, receptor type, J | 12463 | -0.165 | -0.4299 | Yes |
| 79 | RFC3 | RFC3 Entrez, Source | replication factor C (activator 1) 3, 38kDa | 12464 | -0.165 | -0.4137 | Yes |
| 80 | ARPC1B | ARPC1B Entrez, Source | actin related protein 2/3 complex, subunit 1B, 41kDa | 12548 | -0.178 | -0.4025 | Yes |
| 81 | ARL6IP5 | ARL6IP5 Entrez, Source | ADP-ribosylation-like factor 6 interacting protein 5 | 12549 | -0.178 | -0.3851 | Yes |
| 82 | TYMS | TYMS Entrez, Source | thymidylate synthetase | 12607 | -0.185 | -0.3712 | Yes |
| 83 | ANXA2 | ANXA2 Entrez, Source | annexin A2 | 12661 | -0.197 | -0.3560 | Yes |
| 84 | CD47 | CD47 Entrez, Source | CD47 molecule | 12700 | -0.205 | -0.3388 | Yes |
| 85 | PTTG1 | PTTG1 Entrez, Source | pituitary tumor-transforming 1 | 12752 | -0.214 | -0.3218 | Yes |
| 86 | BUB1B | BUB1B Entrez, Source | BUB1 budding uninhibited by benzimidazoles 1 homolog beta (yeast) | 12774 | -0.218 | -0.3020 | Yes |
| 87 | KIF2C | KIF2C Entrez, Source | kinesin family member 2C | 12815 | -0.228 | -0.2827 | Yes |
| 88 | ETS1 | ETS1 Entrez, Source | v-ets erythroblastosis virus E26 oncogene homolog 1 (avian) | 12930 | -0.262 | -0.2657 | Yes |
| 89 | CCNB2 | CCNB2 Entrez, Source | cyclin B2 | 12949 | -0.272 | -0.2404 | Yes |
| 90 | TUBB | TUBB Entrez, Source | tubulin, beta | 13001 | -0.292 | -0.2157 | Yes |
| 91 | FST | FST Entrez, Source | follistatin | 13045 | -0.318 | -0.1879 | Yes |
| 92 | EMP1 | EMP1 Entrez, Source | epithelial membrane protein 1 | 13109 | -0.363 | -0.1571 | Yes |
| 93 | CCNB1 | CCNB1 Entrez, Source | cyclin B1 | 13131 | -0.385 | -0.1211 | Yes |
| 94 | FHL2 | FHL2 Entrez, Source | four and a half LIM domains 2 | 13229 | -0.509 | -0.0786 | Yes |
| 95 | CCNA2 | CCNA2 Entrez, Source | cyclin A2 | 13314 | -0.890 | 0.0021 | Yes |