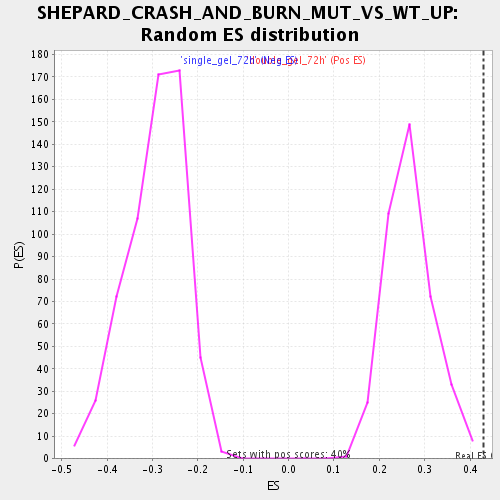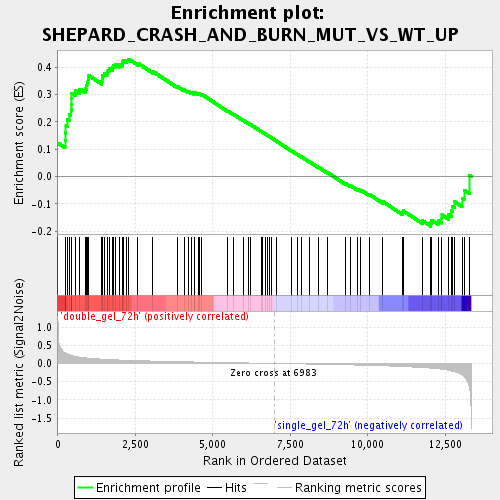
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_72h_versus_single_gel_72h.class.cls #double_gel_72h_versus_single_gel_72h_repos |
| Phenotype | class.cls#double_gel_72h_versus_single_gel_72h_repos |
| Upregulated in class | double_gel_72h |
| GeneSet | SHEPARD_CRASH_AND_BURN_MUT_VS_WT_UP |
| Enrichment Score (ES) | 0.4291969 |
| Normalized Enrichment Score (NES) | 1.6135862 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.063033566 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | FOS | FOS Entrez, Source | v-fos FBJ murine osteosarcoma viral oncogene homolog | 0 | 1.284 | 0.1225 | Yes |
| 2 | RBP4 | RBP4 Entrez, Source | retinol binding protein 4, plasma | 229 | 0.294 | 0.1334 | Yes |
| 3 | C6 | C6 Entrez, Source | complement component 6 | 247 | 0.288 | 0.1595 | Yes |
| 4 | AKR7A2 | AKR7A2 Entrez, Source | aldo-keto reductase family 7, member A2 (aflatoxin aldehyde reductase) | 260 | 0.283 | 0.1857 | Yes |
| 5 | GPD1 | GPD1 Entrez, Source | glycerol-3-phosphate dehydrogenase 1 (soluble) | 296 | 0.267 | 0.2085 | Yes |
| 6 | CYP51A1 | CYP51A1 Entrez, Source | cytochrome P450, family 51, subfamily A, polypeptide 1 | 366 | 0.244 | 0.2265 | Yes |
| 7 | SLC16A6 | SLC16A6 Entrez, Source | solute carrier family 16, member 6 (monocarboxylic acid transporter 7) | 422 | 0.224 | 0.2438 | Yes |
| 8 | EGR1 | EGR1 Entrez, Source | early growth response 1 | 437 | 0.219 | 0.2636 | Yes |
| 9 | AK3L1 | AK3L1 Entrez, Source | adenylate kinase 3-like 1 | 442 | 0.218 | 0.2841 | Yes |
| 10 | CTH | CTH Entrez, Source | cystathionase (cystathionine gamma-lyase) | 447 | 0.216 | 0.3045 | Yes |
| 11 | CYP26A1 | CYP26A1 Entrez, Source | cytochrome P450, family 26, subfamily A, polypeptide 1 | 562 | 0.191 | 0.3141 | Yes |
| 12 | IGFBP1 | IGFBP1 Entrez, Source | insulin-like growth factor binding protein 1 | 703 | 0.173 | 0.3200 | Yes |
| 13 | CEBPB | CEBPB Entrez, Source | CCAAT/enhancer binding protein (C/EBP), beta | 884 | 0.154 | 0.3211 | Yes |
| 14 | PPAN | PPAN Entrez, Source | peter pan homolog (Drosophila) | 924 | 0.151 | 0.3326 | Yes |
| 15 | SHMT1 | SHMT1 Entrez, Source | serine hydroxymethyltransferase 1 (soluble) | 939 | 0.150 | 0.3458 | Yes |
| 16 | DNAJA3 | DNAJA3 Entrez, Source | DnaJ (Hsp40) homolog, subfamily A, member 3 | 972 | 0.147 | 0.3574 | Yes |
| 17 | STARD10 | STARD10 Entrez, Source | START domain containing 10 | 999 | 0.145 | 0.3692 | Yes |
| 18 | ATF3 | ATF3 Entrez, Source | activating transcription factor 3 | 1413 | 0.118 | 0.3493 | Yes |
| 19 | RGC32 | RGC32 Entrez, Source | - | 1424 | 0.117 | 0.3598 | Yes |
| 20 | SSR1 | SSR1 Entrez, Source | signal sequence receptor, alpha (translocon-associated protein alpha) | 1437 | 0.116 | 0.3700 | Yes |
| 21 | JUNB | JUNB Entrez, Source | jun B proto-oncogene | 1487 | 0.114 | 0.3771 | Yes |
| 22 | IMP4 | IMP4 Entrez, Source | IMP4, U3 small nucleolar ribonucleoprotein, homolog (yeast) | 1585 | 0.109 | 0.3803 | Yes |
| 23 | WDR12 | WDR12 Entrez, Source | WD repeat domain 12 | 1615 | 0.108 | 0.3884 | Yes |
| 24 | MRPL12 | MRPL12 Entrez, Source | mitochondrial ribosomal protein L12 | 1652 | 0.107 | 0.3958 | Yes |
| 25 | MKNK2 | MKNK2 Entrez, Source | MAP kinase interacting serine/threonine kinase 2 | 1755 | 0.103 | 0.3980 | Yes |
| 26 | PTGS2 | PTGS2 Entrez, Source | prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) | 1781 | 0.102 | 0.4058 | Yes |
| 27 | GADD45B | GADD45B Entrez, Source | growth arrest and DNA-damage-inducible, beta | 1856 | 0.099 | 0.4097 | Yes |
| 28 | BOP1 | BOP1 Entrez, Source | block of proliferation 1 | 1983 | 0.094 | 0.4092 | Yes |
| 29 | MYBBP1A | MYBBP1A Entrez, Source | MYB binding protein (P160) 1a | 2070 | 0.092 | 0.4114 | Yes |
| 30 | NOG | NOG Entrez, Source | noggin | 2077 | 0.091 | 0.4197 | Yes |
| 31 | MRPS18B | MRPS18B Entrez, Source | mitochondrial ribosomal protein S18B | 2118 | 0.090 | 0.4253 | Yes |
| 32 | DUSP5 | DUSP5 Entrez, Source | dual specificity phosphatase 5 | 2225 | 0.087 | 0.4256 | Yes |
| 33 | HES1 | HES1 Entrez, Source | hairy and enhancer of split 1, (Drosophila) | 2287 | 0.085 | 0.4292 | Yes |
| 34 | PDLIM3 | PDLIM3 Entrez, Source | PDZ and LIM domain 3 | 2579 | 0.077 | 0.4146 | No |
| 35 | CIRH1A | CIRH1A Entrez, Source | cirrhosis, autosomal recessive 1A (cirhin) | 3065 | 0.065 | 0.3842 | No |
| 36 | MMP9 | MMP9 Entrez, Source | matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) | 3861 | 0.049 | 0.3290 | No |
| 37 | PLK3 | PLK3 Entrez, Source | polo-like kinase 3 (Drosophila) | 4077 | 0.045 | 0.3170 | No |
| 38 | CYP27B1 | CYP27B1 Entrez, Source | cytochrome P450, family 27, subfamily B, polypeptide 1 | 4218 | 0.043 | 0.3105 | No |
| 39 | FTSJ3 | FTSJ3 Entrez, Source | FtsJ homolog 3 (E. coli) | 4307 | 0.041 | 0.3078 | No |
| 40 | CORT | CORT Entrez, Source | cortistatin | 4395 | 0.040 | 0.3050 | No |
| 41 | ETF1 | ETF1 Entrez, Source | eukaryotic translation termination factor 1 | 4420 | 0.039 | 0.3070 | No |
| 42 | PFKFB3 | PFKFB3 Entrez, Source | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 | 4523 | 0.037 | 0.3028 | No |
| 43 | DUSP4 | DUSP4 Entrez, Source | dual specificity phosphatase 4 | 4576 | 0.037 | 0.3024 | No |
| 44 | BTG2 | BTG2 Entrez, Source | BTG family, member 2 | 4648 | 0.035 | 0.3004 | No |
| 45 | ODC1 | ODC1 Entrez, Source | ornithine decarboxylase 1 | 5487 | 0.022 | 0.2393 | No |
| 46 | FBL | FBL Entrez, Source | fibrillarin | 5673 | 0.019 | 0.2272 | No |
| 47 | AP3M1 | AP3M1 Entrez, Source | adaptor-related protein complex 3, mu 1 subunit | 5986 | 0.014 | 0.2050 | No |
| 48 | TP53 | TP53 Entrez, Source | tumor protein p53 (Li-Fraumeni syndrome) | 6158 | 0.012 | 0.1932 | No |
| 49 | PIM1 | PIM1 Entrez, Source | pim-1 oncogene | 6208 | 0.011 | 0.1906 | No |
| 50 | TOB1 | TOB1 Entrez, Source | transducer of ERBB2, 1 | 6567 | 0.006 | 0.1642 | No |
| 51 | IGF2 | IGF2 Entrez, Source | insulin-like growth factor 2 (somatomedin A) | 6610 | 0.005 | 0.1615 | No |
| 52 | EIF4E | EIF4E Entrez, Source | eukaryotic translation initiation factor 4E | 6703 | 0.004 | 0.1549 | No |
| 53 | ID2 | ID2 Entrez, Source | inhibitor of DNA binding 2, dominant negative helix-loop-helix protein | 6773 | 0.003 | 0.1500 | No |
| 54 | SLC3A2 | SLC3A2 Entrez, Source | solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2 | 6817 | 0.002 | 0.1470 | No |
| 55 | SRPRB | SRPRB Entrez, Source | signal recognition particle receptor, B subunit | 6895 | 0.001 | 0.1413 | No |
| 56 | TOMM20 | TOMM20 Entrez, Source | translocase of outer mitochondrial membrane 20 homolog (yeast) | 7046 | -0.001 | 0.1301 | No |
| 57 | TAF9 | TAF9 Entrez, Source | TAF9 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 32kDa | 7531 | -0.008 | 0.0943 | No |
| 58 | POP4 | POP4 Entrez, Source | processing of precursor 4, ribonuclease P/MRP subunit (S. cerevisiae) | 7726 | -0.011 | 0.0807 | No |
| 59 | MYOG | MYOG Entrez, Source | myogenin (myogenic factor 4) | 7858 | -0.013 | 0.0721 | No |
| 60 | RASL11B | RASL11B Entrez, Source | RAS-like, family 11, member B | 8125 | -0.017 | 0.0536 | No |
| 61 | ATPIF1 | ATPIF1 Entrez, Source | ATPase inhibitory factor 1 | 8423 | -0.022 | 0.0333 | No |
| 62 | SLC12A3 | SLC12A3 Entrez, Source | solute carrier family 12 (sodium/chloride transporters), member 3 | 8709 | -0.026 | 0.0143 | No |
| 63 | ITGB4BP | ITGB4BP Entrez, Source | integrin beta 4 binding protein | 9293 | -0.038 | -0.0261 | No |
| 64 | SNAI1 | SNAI1 Entrez, Source | snail homolog 1 (Drosophila) | 9442 | -0.040 | -0.0334 | No |
| 65 | MRPS18A | MRPS18A Entrez, Source | mitochondrial ribosomal protein S18A | 9675 | -0.045 | -0.0466 | No |
| 66 | BAX | BAX Entrez, Source | BCL2-associated X protein | 9773 | -0.047 | -0.0494 | No |
| 67 | FN1 | FN1 Entrez, Source | fibronectin 1 | 10062 | -0.053 | -0.0660 | No |
| 68 | CLDN4 | CLDN4 Entrez, Source | claudin 4 | 10487 | -0.064 | -0.0919 | No |
| 69 | PPP1R14A | PPP1R14A Entrez, Source | protein phosphatase 1, regulatory (inhibitor) subunit 14A | 11121 | -0.083 | -0.1318 | No |
| 70 | CCNG1 | CCNG1 Entrez, Source | cyclin G1 | 11138 | -0.083 | -0.1250 | No |
| 71 | JUN | JUN Entrez, Source | jun oncogene | 11756 | -0.111 | -0.1609 | No |
| 72 | DSCR1 | DSCR1 Entrez, Source | Down syndrome critical region gene 1 | 12036 | -0.127 | -0.1698 | No |
| 73 | ADAM10 | ADAM10 Entrez, Source | ADAM metallopeptidase domain 10 | 12061 | -0.129 | -0.1594 | No |
| 74 | CYR61 | CYR61 Entrez, Source | cysteine-rich, angiogenic inducer, 61 | 12269 | -0.146 | -0.1611 | No |
| 75 | SLC12A7 | SLC12A7 Entrez, Source | solute carrier family 12 (potassium/chloride transporters), member 7 | 12391 | -0.157 | -0.1553 | No |
| 76 | CASP8 | CASP8 Entrez, Source | caspase 8, apoptosis-related cysteine peptidase | 12395 | -0.157 | -0.1405 | No |
| 77 | MMP2 | MMP2 Entrez, Source | matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase) | 12615 | -0.187 | -0.1392 | No |
| 78 | TXNIP | TXNIP Entrez, Source | thioredoxin interacting protein | 12699 | -0.205 | -0.1259 | No |
| 79 | SGK | SGK Entrez, Source | serum/glucocorticoid regulated kinase | 12735 | -0.210 | -0.1085 | No |
| 80 | BMP2 | BMP2 Entrez, Source | bone morphogenetic protein 2 | 12792 | -0.222 | -0.0915 | No |
| 81 | FST | FST Entrez, Source | follistatin | 13045 | -0.318 | -0.0802 | No |
| 82 | HSPA4 | HSPA4 Entrez, Source | heat shock 70kDa protein 4 | 13114 | -0.366 | -0.0504 | No |
| 83 | SERPINH1 | SERPINH1 Entrez, Source | serpin peptidase inhibitor, clade H (heat shock protein 47), member 1, (collagen binding protein 1) | 13290 | -0.708 | 0.0039 | No |