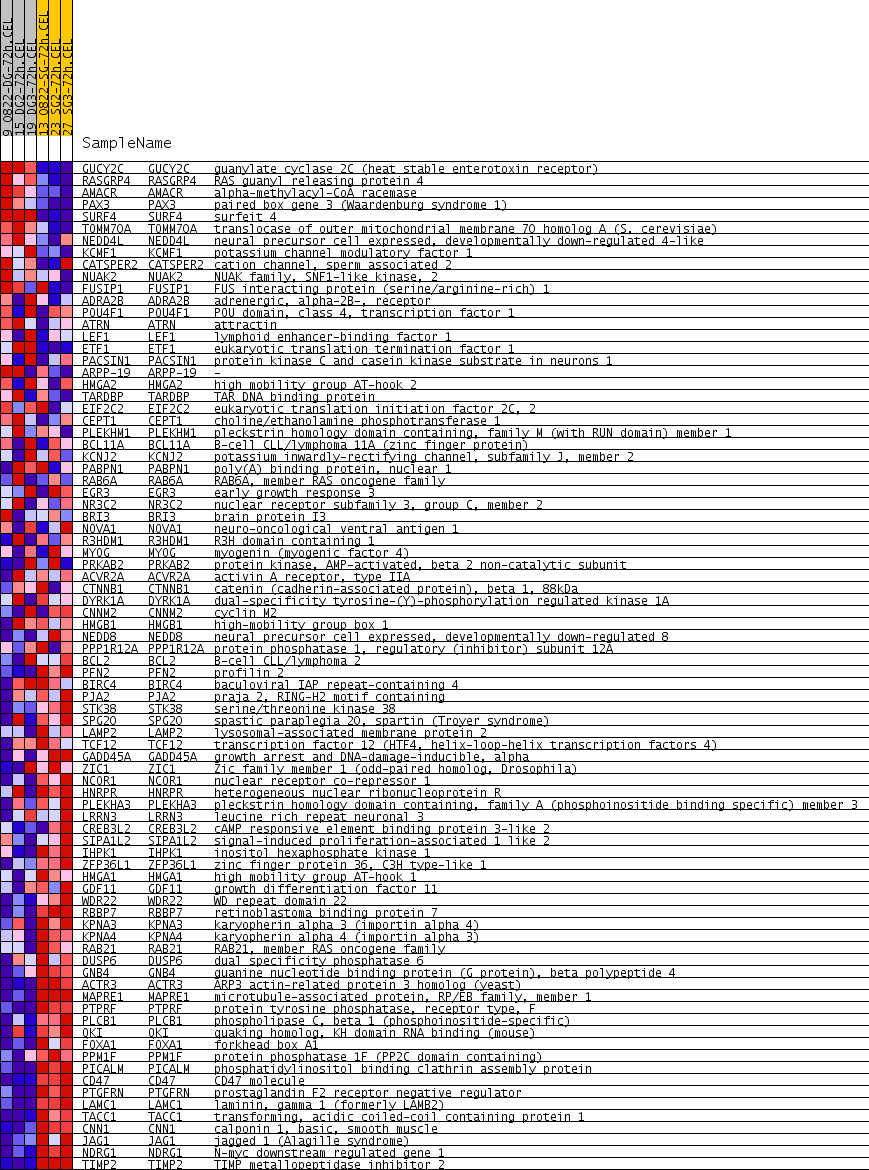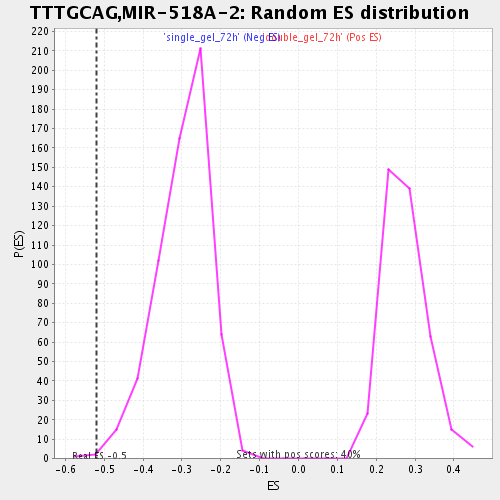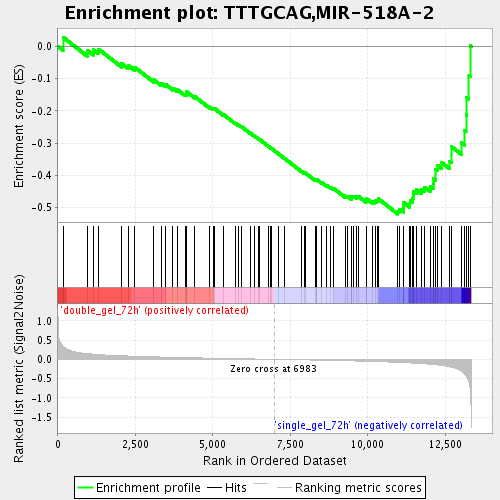
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_72h_versus_single_gel_72h.class.cls #double_gel_72h_versus_single_gel_72h_repos |
| Phenotype | class.cls#double_gel_72h_versus_single_gel_72h_repos |
| Upregulated in class | single_gel_72h |
| GeneSet | TTTGCAG,MIR-518A-2 |
| Enrichment Score (ES) | -0.520122 |
| Normalized Enrichment Score (NES) | -1.7526215 |
| Nominal p-value | 0.0016528926 |
| FDR q-value | 0.047038477 |
| FWER p-Value | 0.999 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | GUCY2C | GUCY2C Entrez, Source | guanylate cyclase 2C (heat stable enterotoxin receptor) | 167 | 0.341 | 0.0287 | No |
| 2 | RASGRP4 | RASGRP4 Entrez, Source | RAS guanyl releasing protein 4 | 946 | 0.149 | -0.0119 | No |
| 3 | AMACR | AMACR Entrez, Source | alpha-methylacyl-CoA racemase | 1143 | 0.134 | -0.0104 | No |
| 4 | PAX3 | PAX3 Entrez, Source | paired box gene 3 (Waardenburg syndrome 1) | 1312 | 0.123 | -0.0081 | No |
| 5 | SURF4 | SURF4 Entrez, Source | surfeit 4 | 2054 | 0.092 | -0.0528 | No |
| 6 | TOMM70A | TOMM70A Entrez, Source | translocase of outer mitochondrial membrane 70 homolog A (S. cerevisiae) | 2268 | 0.086 | -0.0585 | No |
| 7 | NEDD4L | NEDD4L Entrez, Source | neural precursor cell expressed, developmentally down-regulated 4-like | 2471 | 0.081 | -0.0639 | No |
| 8 | KCMF1 | KCMF1 Entrez, Source | potassium channel modulatory factor 1 | 3093 | 0.064 | -0.1029 | No |
| 9 | CATSPER2 | CATSPER2 Entrez, Source | cation channel, sperm associated 2 | 3345 | 0.059 | -0.1147 | No |
| 10 | NUAK2 | NUAK2 Entrez, Source | NUAK family, SNF1-like kinase, 2 | 3483 | 0.056 | -0.1182 | No |
| 11 | FUSIP1 | FUSIP1 Entrez, Source | FUS interacting protein (serine/arginine-rich) 1 | 3709 | 0.052 | -0.1289 | No |
| 12 | ADRA2B | ADRA2B Entrez, Source | adrenergic, alpha-2B-, receptor | 3846 | 0.049 | -0.1332 | No |
| 13 | POU4F1 | POU4F1 Entrez, Source | POU domain, class 4, transcription factor 1 | 4112 | 0.044 | -0.1478 | No |
| 14 | ATRN | ATRN Entrez, Source | attractin | 4158 | 0.044 | -0.1459 | No |
| 15 | LEF1 | LEF1 Entrez, Source | lymphoid enhancer-binding factor 1 | 4161 | 0.044 | -0.1408 | No |
| 16 | ETF1 | ETF1 Entrez, Source | eukaryotic translation termination factor 1 | 4420 | 0.039 | -0.1555 | No |
| 17 | PACSIN1 | PACSIN1 Entrez, Source | protein kinase C and casein kinase substrate in neurons 1 | 4894 | 0.031 | -0.1874 | No |
| 18 | ARPP-19 | ARPP-19 Entrez, Source | - | 5020 | 0.029 | -0.1934 | No |
| 19 | HMGA2 | HMGA2 Entrez, Source | high mobility group AT-hook 2 | 5052 | 0.028 | -0.1923 | No |
| 20 | TARDBP | TARDBP Entrez, Source | TAR DNA binding protein | 5332 | 0.024 | -0.2104 | No |
| 21 | EIF2C2 | EIF2C2 Entrez, Source | eukaryotic translation initiation factor 2C, 2 | 5725 | 0.018 | -0.2377 | No |
| 22 | CEPT1 | CEPT1 Entrez, Source | choline/ethanolamine phosphotransferase 1 | 5830 | 0.017 | -0.2435 | No |
| 23 | PLEKHM1 | PLEKHM1 Entrez, Source | pleckstrin homology domain containing, family M (with RUN domain) member 1 | 5913 | 0.015 | -0.2478 | No |
| 24 | BCL11A | BCL11A Entrez, Source | B-cell CLL/lymphoma 11A (zinc finger protein) | 6200 | 0.011 | -0.2680 | No |
| 25 | KCNJ2 | KCNJ2 Entrez, Source | potassium inwardly-rectifying channel, subfamily J, member 2 | 6345 | 0.009 | -0.2778 | No |
| 26 | PABPN1 | PABPN1 Entrez, Source | poly(A) binding protein, nuclear 1 | 6488 | 0.007 | -0.2877 | No |
| 27 | RAB6A | RAB6A Entrez, Source | RAB6A, member RAS oncogene family | 6512 | 0.006 | -0.2887 | No |
| 28 | EGR3 | EGR3 Entrez, Source | early growth response 3 | 6801 | 0.003 | -0.3101 | No |
| 29 | NR3C2 | NR3C2 Entrez, Source | nuclear receptor subfamily 3, group C, member 2 | 6857 | 0.002 | -0.3140 | No |
| 30 | BRI3 | BRI3 Entrez, Source | brain protein I3 | 6885 | 0.002 | -0.3159 | No |
| 31 | NOVA1 | NOVA1 Entrez, Source | neuro-oncological ventral antigen 1 | 7115 | -0.002 | -0.3329 | No |
| 32 | R3HDM1 | R3HDM1 Entrez, Source | R3H domain containing 1 | 7305 | -0.005 | -0.3466 | No |
| 33 | MYOG | MYOG Entrez, Source | myogenin (myogenic factor 4) | 7858 | -0.013 | -0.3866 | No |
| 34 | PRKAB2 | PRKAB2 Entrez, Source | protein kinase, AMP-activated, beta 2 non-catalytic subunit | 7945 | -0.014 | -0.3914 | No |
| 35 | ACVR2A | ACVR2A Entrez, Source | activin A receptor, type IIA | 7984 | -0.015 | -0.3925 | No |
| 36 | CTNNB1 | CTNNB1 Entrez, Source | catenin (cadherin-associated protein), beta 1, 88kDa | 8312 | -0.020 | -0.4147 | No |
| 37 | DYRK1A | DYRK1A Entrez, Source | dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A | 8340 | -0.020 | -0.4143 | No |
| 38 | CNNM2 | CNNM2 Entrez, Source | cyclin M2 | 8352 | -0.021 | -0.4126 | No |
| 39 | HMGB1 | HMGB1 Entrez, Source | high-mobility group box 1 | 8508 | -0.023 | -0.4215 | No |
| 40 | NEDD8 | NEDD8 Entrez, Source | neural precursor cell expressed, developmentally down-regulated 8 | 8682 | -0.026 | -0.4314 | No |
| 41 | PPP1R12A | PPP1R12A Entrez, Source | protein phosphatase 1, regulatory (inhibitor) subunit 12A | 8798 | -0.028 | -0.4366 | No |
| 42 | BCL2 | BCL2 Entrez, Source | B-cell CLL/lymphoma 2 | 8887 | -0.030 | -0.4397 | No |
| 43 | PFN2 | PFN2 Entrez, Source | profilin 2 | 9269 | -0.037 | -0.4639 | No |
| 44 | BIRC4 | BIRC4 Entrez, Source | baculoviral IAP repeat-containing 4 | 9335 | -0.038 | -0.4641 | No |
| 45 | PJA2 | PJA2 Entrez, Source | praja 2, RING-H2 motif containing | 9485 | -0.041 | -0.4703 | No |
| 46 | STK38 | STK38 Entrez, Source | serine/threonine kinase 38 | 9486 | -0.041 | -0.4653 | No |
| 47 | SPG20 | SPG20 Entrez, Source | spastic paraplegia 20, spartin (Troyer syndrome) | 9547 | -0.043 | -0.4647 | No |
| 48 | LAMP2 | LAMP2 Entrez, Source | lysosomal-associated membrane protein 2 | 9636 | -0.044 | -0.4660 | No |
| 49 | TCF12 | TCF12 Entrez, Source | transcription factor 12 (HTF4, helix-loop-helix transcription factors 4) | 9690 | -0.045 | -0.4644 | No |
| 50 | GADD45A | GADD45A Entrez, Source | growth arrest and DNA-damage-inducible, alpha | 9946 | -0.051 | -0.4775 | No |
| 51 | ZIC1 | ZIC1 Entrez, Source | Zic family member 1 (odd-paired homolog, Drosophila) | 9968 | -0.051 | -0.4728 | No |
| 52 | NCOR1 | NCOR1 Entrez, Source | nuclear receptor co-repressor 1 | 10165 | -0.056 | -0.4808 | No |
| 53 | HNRPR | HNRPR Entrez, Source | heterogeneous nuclear ribonucleoprotein R | 10233 | -0.058 | -0.4789 | No |
| 54 | PLEKHA3 | PLEKHA3 Entrez, Source | pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 3 | 10298 | -0.059 | -0.4766 | No |
| 55 | LRRN3 | LRRN3 Entrez, Source | leucine rich repeat neuronal 3 | 10341 | -0.060 | -0.4725 | No |
| 56 | CREB3L2 | CREB3L2 Entrez, Source | cAMP responsive element binding protein 3-like 2 | 10974 | -0.078 | -0.5107 | Yes |
| 57 | SIPA1L2 | SIPA1L2 Entrez, Source | signal-induced proliferation-associated 1 like 2 | 11018 | -0.079 | -0.5043 | Yes |
| 58 | IHPK1 | IHPK1 Entrez, Source | inositol hexaphosphate kinase 1 | 11145 | -0.083 | -0.5037 | Yes |
| 59 | ZFP36L1 | ZFP36L1 Entrez, Source | zinc finger protein 36, C3H type-like 1 | 11158 | -0.084 | -0.4944 | Yes |
| 60 | HMGA1 | HMGA1 Entrez, Source | high mobility group AT-hook 1 | 11163 | -0.084 | -0.4845 | Yes |
| 61 | GDF11 | GDF11 Entrez, Source | growth differentiation factor 11 | 11359 | -0.093 | -0.4879 | Yes |
| 62 | WDR22 | WDR22 Entrez, Source | WD repeat domain 22 | 11366 | -0.093 | -0.4771 | Yes |
| 63 | RBBP7 | RBBP7 Entrez, Source | retinoblastoma binding protein 7 | 11453 | -0.098 | -0.4717 | Yes |
| 64 | KPNA3 | KPNA3 Entrez, Source | karyopherin alpha 3 (importin alpha 4) | 11464 | -0.098 | -0.4606 | Yes |
| 65 | KPNA4 | KPNA4 Entrez, Source | karyopherin alpha 4 (importin alpha 3) | 11478 | -0.099 | -0.4496 | Yes |
| 66 | RAB21 | RAB21 Entrez, Source | RAB21, member RAS oncogene family | 11573 | -0.103 | -0.4442 | Yes |
| 67 | DUSP6 | DUSP6 Entrez, Source | dual specificity phosphatase 6 | 11745 | -0.111 | -0.4436 | Yes |
| 68 | GNB4 | GNB4 Entrez, Source | guanine nucleotide binding protein (G protein), beta polypeptide 4 | 11832 | -0.115 | -0.4361 | Yes |
| 69 | ACTR3 | ACTR3 Entrez, Source | ARP3 actin-related protein 3 homolog (yeast) | 12015 | -0.126 | -0.4346 | Yes |
| 70 | MAPRE1 | MAPRE1 Entrez, Source | microtubule-associated protein, RP/EB family, member 1 | 12109 | -0.132 | -0.4256 | Yes |
| 71 | PTPRF | PTPRF Entrez, Source | protein tyrosine phosphatase, receptor type, F | 12127 | -0.133 | -0.4107 | Yes |
| 72 | PLCB1 | PLCB1 Entrez, Source | phospholipase C, beta 1 (phosphoinositide-specific) | 12183 | -0.138 | -0.3982 | Yes |
| 73 | QKI | QKI Entrez, Source | quaking homolog, KH domain RNA binding (mouse) | 12184 | -0.138 | -0.3815 | Yes |
| 74 | FOXA1 | FOXA1 Entrez, Source | forkhead box A1 | 12239 | -0.143 | -0.3682 | Yes |
| 75 | PPM1F | PPM1F Entrez, Source | protein phosphatase 1F (PP2C domain containing) | 12392 | -0.157 | -0.3606 | Yes |
| 76 | PICALM | PICALM Entrez, Source | phosphatidylinositol binding clathrin assembly protein | 12640 | -0.193 | -0.3559 | Yes |
| 77 | CD47 | CD47 Entrez, Source | CD47 molecule | 12700 | -0.205 | -0.3355 | Yes |
| 78 | PTGFRN | PTGFRN Entrez, Source | prostaglandin F2 receptor negative regulator | 12701 | -0.205 | -0.3106 | Yes |
| 79 | LAMC1 | LAMC1 Entrez, Source | laminin, gamma 1 (formerly LAMB2) | 13018 | -0.300 | -0.2981 | Yes |
| 80 | TACC1 | TACC1 Entrez, Source | transforming, acidic coiled-coil containing protein 1 | 13123 | -0.377 | -0.2602 | Yes |
| 81 | CNN1 | CNN1 Entrez, Source | calponin 1, basic, smooth muscle | 13171 | -0.429 | -0.2118 | Yes |
| 82 | JAG1 | JAG1 Entrez, Source | jagged 1 (Alagille syndrome) | 13192 | -0.458 | -0.1578 | Yes |
| 83 | NDRG1 | NDRG1 Entrez, Source | N-myc downstream regulated gene 1 | 13266 | -0.607 | -0.0897 | Yes |
| 84 | TIMP2 | TIMP2 Entrez, Source | TIMP metallopeptidase inhibitor 2 | 13306 | -0.786 | 0.0027 | Yes |