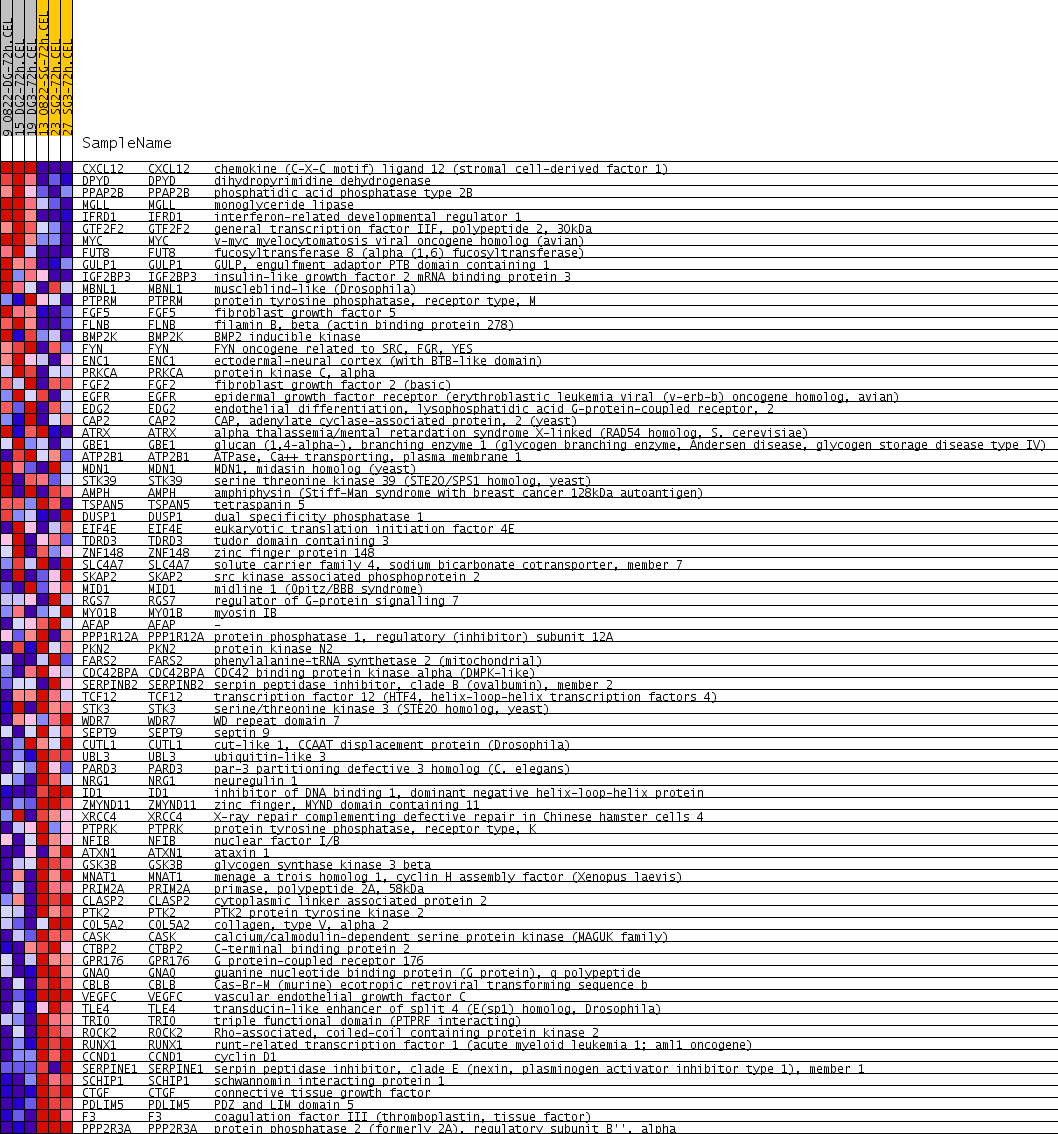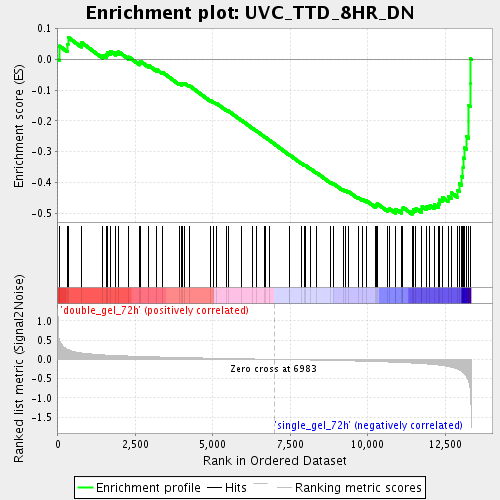
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_72h_versus_single_gel_72h.class.cls #double_gel_72h_versus_single_gel_72h_repos |
| Phenotype | class.cls#double_gel_72h_versus_single_gel_72h_repos |
| Upregulated in class | single_gel_72h |
| GeneSet | UVC_TTD_8HR_DN |
| Enrichment Score (ES) | -0.50314164 |
| Normalized Enrichment Score (NES) | -1.7313453 |
| Nominal p-value | 0.0033167496 |
| FDR q-value | 0.052143533 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | CXCL12 | CXCL12 Entrez, Source | chemokine (C-X-C motif) ligand 12 (stromal cell-derived factor 1) | 50 | 0.500 | 0.0433 | No |
| 2 | DPYD | DPYD Entrez, Source | dihydropyrimidine dehydrogenase | 303 | 0.265 | 0.0492 | No |
| 3 | PPAP2B | PPAP2B Entrez, Source | phosphatidic acid phosphatase type 2B | 329 | 0.257 | 0.0716 | No |
| 4 | MGLL | MGLL Entrez, Source | monoglyceride lipase | 754 | 0.167 | 0.0553 | No |
| 5 | IFRD1 | IFRD1 Entrez, Source | interferon-related developmental regulator 1 | 1449 | 0.116 | 0.0139 | No |
| 6 | GTF2F2 | GTF2F2 Entrez, Source | general transcription factor IIF, polypeptide 2, 30kDa | 1569 | 0.110 | 0.0152 | No |
| 7 | MYC | MYC Entrez, Source | v-myc myelocytomatosis viral oncogene homolog (avian) | 1609 | 0.108 | 0.0225 | No |
| 8 | FUT8 | FUT8 Entrez, Source | fucosyltransferase 8 (alpha (1,6) fucosyltransferase) | 1701 | 0.105 | 0.0255 | No |
| 9 | GULP1 | GULP1 Entrez, Source | GULP, engulfment adaptor PTB domain containing 1 | 1862 | 0.099 | 0.0228 | No |
| 10 | IGF2BP3 | IGF2BP3 Entrez, Source | insulin-like growth factor 2 mRNA binding protein 3 | 1965 | 0.095 | 0.0240 | No |
| 11 | MBNL1 | MBNL1 Entrez, Source | muscleblind-like (Drosophila) | 2291 | 0.085 | 0.0075 | No |
| 12 | PTPRM | PTPRM Entrez, Source | protein tyrosine phosphatase, receptor type, M | 2639 | 0.075 | -0.0116 | No |
| 13 | FGF5 | FGF5 Entrez, Source | fibroblast growth factor 5 | 2656 | 0.075 | -0.0057 | No |
| 14 | FLNB | FLNB Entrez, Source | filamin B, beta (actin binding protein 278) | 2930 | 0.068 | -0.0199 | No |
| 15 | BMP2K | BMP2K Entrez, Source | BMP2 inducible kinase | 3182 | 0.063 | -0.0330 | No |
| 16 | FYN | FYN Entrez, Source | FYN oncogene related to SRC, FGR, YES | 3374 | 0.059 | -0.0418 | No |
| 17 | ENC1 | ENC1 Entrez, Source | ectodermal-neural cortex (with BTB-like domain) | 3917 | 0.048 | -0.0782 | No |
| 18 | PRKCA | PRKCA Entrez, Source | protein kinase C, alpha | 3999 | 0.046 | -0.0799 | No |
| 19 | FGF2 | FGF2 Entrez, Source | fibroblast growth factor 2 (basic) | 4030 | 0.046 | -0.0779 | No |
| 20 | EGFR | EGFR Entrez, Source | epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | 4079 | 0.045 | -0.0773 | No |
| 21 | EDG2 | EDG2 Entrez, Source | endothelial differentiation, lysophosphatidic acid G-protein-coupled receptor, 2 | 4234 | 0.042 | -0.0849 | No |
| 22 | CAP2 | CAP2 Entrez, Source | CAP, adenylate cyclase-associated protein, 2 (yeast) | 4920 | 0.030 | -0.1337 | No |
| 23 | ATRX | ATRX Entrez, Source | alpha thalassemia/mental retardation syndrome X-linked (RAD54 homolog, S. cerevisiae) | 5030 | 0.029 | -0.1392 | No |
| 24 | GBE1 | GBE1 Entrez, Source | glucan (1,4-alpha-), branching enzyme 1 (glycogen branching enzyme, Andersen disease, glycogen storage disease type IV) | 5119 | 0.027 | -0.1433 | No |
| 25 | ATP2B1 | ATP2B1 Entrez, Source | ATPase, Ca++ transporting, plasma membrane 1 | 5439 | 0.023 | -0.1652 | No |
| 26 | MDN1 | MDN1 Entrez, Source | MDN1, midasin homolog (yeast) | 5490 | 0.022 | -0.1669 | No |
| 27 | STK39 | STK39 Entrez, Source | serine threonine kinase 39 (STE20/SPS1 homolog, yeast) | 5922 | 0.015 | -0.1980 | No |
| 28 | AMPH | AMPH Entrez, Source | amphiphysin (Stiff-Man syndrome with breast cancer 128kDa autoantigen) | 6291 | 0.010 | -0.2248 | No |
| 29 | TSPAN5 | TSPAN5 Entrez, Source | tetraspanin 5 | 6410 | 0.008 | -0.2330 | No |
| 30 | DUSP1 | DUSP1 Entrez, Source | dual specificity phosphatase 1 | 6673 | 0.004 | -0.2523 | No |
| 31 | EIF4E | EIF4E Entrez, Source | eukaryotic translation initiation factor 4E | 6703 | 0.004 | -0.2542 | No |
| 32 | TDRD3 | TDRD3 Entrez, Source | tudor domain containing 3 | 6836 | 0.002 | -0.2639 | No |
| 33 | ZNF148 | ZNF148 Entrez, Source | zinc finger protein 148 | 7480 | -0.007 | -0.3117 | No |
| 34 | SLC4A7 | SLC4A7 Entrez, Source | solute carrier family 4, sodium bicarbonate cotransporter, member 7 | 7486 | -0.007 | -0.3114 | No |
| 35 | SKAP2 | SKAP2 Entrez, Source | src kinase associated phosphoprotein 2 | 7850 | -0.013 | -0.3376 | No |
| 36 | MID1 | MID1 Entrez, Source | midline 1 (Opitz/BBB syndrome) | 7962 | -0.014 | -0.3446 | No |
| 37 | RGS7 | RGS7 Entrez, Source | regulator of G-protein signalling 7 | 7991 | -0.015 | -0.3453 | No |
| 38 | MYO1B | MYO1B Entrez, Source | myosin IB | 8141 | -0.017 | -0.3550 | No |
| 39 | AFAP | AFAP Entrez, Source | - | 8351 | -0.021 | -0.3688 | No |
| 40 | PPP1R12A | PPP1R12A Entrez, Source | protein phosphatase 1, regulatory (inhibitor) subunit 12A | 8798 | -0.028 | -0.3998 | No |
| 41 | PKN2 | PKN2 Entrez, Source | protein kinase N2 | 8882 | -0.030 | -0.4032 | No |
| 42 | FARS2 | FARS2 Entrez, Source | phenylalanine-tRNA synthetase 2 (mitochondrial) | 9209 | -0.036 | -0.4244 | No |
| 43 | CDC42BPA | CDC42BPA Entrez, Source | CDC42 binding protein kinase alpha (DMPK-like) | 9266 | -0.037 | -0.4252 | No |
| 44 | SERPINB2 | SERPINB2 Entrez, Source | serpin peptidase inhibitor, clade B (ovalbumin), member 2 | 9372 | -0.039 | -0.4294 | No |
| 45 | TCF12 | TCF12 Entrez, Source | transcription factor 12 (HTF4, helix-loop-helix transcription factors 4) | 9690 | -0.045 | -0.4490 | No |
| 46 | STK3 | STK3 Entrez, Source | serine/threonine kinase 3 (STE20 homolog, yeast) | 9831 | -0.048 | -0.4550 | No |
| 47 | WDR7 | WDR7 Entrez, Source | WD repeat domain 7 | 9952 | -0.051 | -0.4593 | No |
| 48 | SEPT9 | SEPT9 Entrez, Source | septin 9 | 10261 | -0.058 | -0.4770 | No |
| 49 | CUTL1 | CUTL1 Entrez, Source | cut-like 1, CCAAT displacement protein (Drosophila) | 10279 | -0.058 | -0.4728 | No |
| 50 | UBL3 | UBL3 Entrez, Source | ubiquitin-like 3 | 10310 | -0.059 | -0.4695 | No |
| 51 | PARD3 | PARD3 Entrez, Source | par-3 partitioning defective 3 homolog (C. elegans) | 10642 | -0.068 | -0.4881 | No |
| 52 | NRG1 | NRG1 Entrez, Source | neuregulin 1 | 10701 | -0.070 | -0.4859 | No |
| 53 | ID1 | ID1 Entrez, Source | inhibitor of DNA binding 1, dominant negative helix-loop-helix protein | 10891 | -0.075 | -0.4931 | No |
| 54 | ZMYND11 | ZMYND11 Entrez, Source | zinc finger, MYND domain containing 11 | 10901 | -0.075 | -0.4867 | No |
| 55 | XRCC4 | XRCC4 Entrez, Source | X-ray repair complementing defective repair in Chinese hamster cells 4 | 11084 | -0.081 | -0.4928 | No |
| 56 | PTPRK | PTPRK Entrez, Source | protein tyrosine phosphatase, receptor type, K | 11105 | -0.082 | -0.4865 | No |
| 57 | NFIB | NFIB Entrez, Source | nuclear factor I/B | 11136 | -0.083 | -0.4810 | No |
| 58 | ATXN1 | ATXN1 Entrez, Source | ataxin 1 | 11431 | -0.096 | -0.4941 | Yes |
| 59 | GSK3B | GSK3B Entrez, Source | glycogen synthase kinase 3 beta | 11474 | -0.098 | -0.4880 | Yes |
| 60 | MNAT1 | MNAT1 Entrez, Source | menage a trois homolog 1, cyclin H assembly factor (Xenopus laevis) | 11555 | -0.102 | -0.4844 | Yes |
| 61 | PRIM2A | PRIM2A Entrez, Source | primase, polypeptide 2A, 58kDa | 11731 | -0.111 | -0.4872 | Yes |
| 62 | CLASP2 | CLASP2 Entrez, Source | cytoplasmic linker associated protein 2 | 11749 | -0.111 | -0.4780 | Yes |
| 63 | PTK2 | PTK2 Entrez, Source | PTK2 protein tyrosine kinase 2 | 11904 | -0.119 | -0.4785 | Yes |
| 64 | COL5A2 | COL5A2 Entrez, Source | collagen, type V, alpha 2 | 12008 | -0.126 | -0.4744 | Yes |
| 65 | CASK | CASK Entrez, Source | calcium/calmodulin-dependent serine protein kinase (MAGUK family) | 12154 | -0.135 | -0.4727 | Yes |
| 66 | CTBP2 | CTBP2 Entrez, Source | C-terminal binding protein 2 | 12294 | -0.147 | -0.4693 | Yes |
| 67 | GPR176 | GPR176 Entrez, Source | G protein-coupled receptor 176 | 12322 | -0.150 | -0.4572 | Yes |
| 68 | GNAQ | GNAQ Entrez, Source | guanine nucleotide binding protein (G protein), q polypeptide | 12403 | -0.158 | -0.4484 | Yes |
| 69 | CBLB | CBLB Entrez, Source | Cas-Br-M (murine) ecotropic retroviral transforming sequence b | 12594 | -0.184 | -0.4454 | Yes |
| 70 | VEGFC | VEGFC Entrez, Source | vascular endothelial growth factor C | 12697 | -0.204 | -0.4339 | Yes |
| 71 | TLE4 | TLE4 Entrez, Source | transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) | 12889 | -0.247 | -0.4250 | Yes |
| 72 | TRIO | TRIO Entrez, Source | triple functional domain (PTPRF interacting) | 12955 | -0.274 | -0.4041 | Yes |
| 73 | ROCK2 | ROCK2 Entrez, Source | Rho-associated, coiled-coil containing protein kinase 2 | 13035 | -0.310 | -0.3809 | Yes |
| 74 | RUNX1 | RUNX1 Entrez, Source | runt-related transcription factor 1 (acute myeloid leukemia 1; aml1 oncogene) | 13073 | -0.336 | -0.3521 | Yes |
| 75 | CCND1 | CCND1 Entrez, Source | cyclin D1 | 13085 | -0.343 | -0.3207 | Yes |
| 76 | SERPINE1 | SERPINE1 Entrez, Source | serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 | 13125 | -0.378 | -0.2881 | Yes |
| 77 | SCHIP1 | SCHIP1 Entrez, Source | schwannomin interacting protein 1 | 13180 | -0.438 | -0.2509 | Yes |
| 78 | CTGF | CTGF Entrez, Source | connective tissue growth factor | 13260 | -0.571 | -0.2031 | Yes |
| 79 | PDLIM5 | PDLIM5 Entrez, Source | PDZ and LIM domain 5 | 13261 | -0.575 | -0.1490 | Yes |
| 80 | F3 | F3 Entrez, Source | coagulation factor III (thromboplastin, tissue factor) | 13308 | -0.800 | -0.0772 | Yes |
| 81 | PPP2R3A | PPP2R3A Entrez, Source | protein phosphatase 2 (formerly 2A), regulatory subunit B'', alpha | 13312 | -0.847 | 0.0023 | Yes |