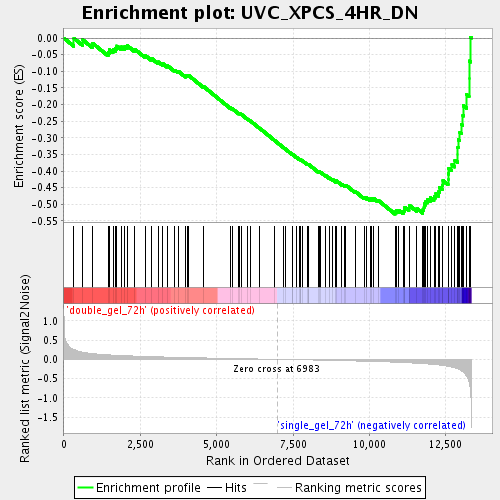
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_72h_versus_single_gel_72h.class.cls #double_gel_72h_versus_single_gel_72h_repos |
| Phenotype | class.cls#double_gel_72h_versus_single_gel_72h_repos |
| Upregulated in class | single_gel_72h |
| GeneSet | UVC_XPCS_4HR_DN |
| Enrichment Score (ES) | -0.5296858 |
| Normalized Enrichment Score (NES) | -1.8462214 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.028396029 |
| FWER p-Value | 0.834 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | MECP2 | MECP2 Entrez, Source | methyl CpG binding protein 2 (Rett syndrome) | 327 | 0.257 | -0.0006 | No |
| 2 | BHLHB2 | BHLHB2 Entrez, Source | basic helix-loop-helix domain containing, class B, 2 | 613 | 0.184 | -0.0049 | No |
| 3 | PRKCBP1 | PRKCBP1 Entrez, Source | protein kinase C binding protein 1 | 932 | 0.150 | -0.0149 | No |
| 4 | IFRD1 | IFRD1 Entrez, Source | interferon-related developmental regulator 1 | 1449 | 0.116 | -0.0430 | No |
| 5 | DLC1 | DLC1 Entrez, Source | deleted in liver cancer 1 | 1483 | 0.114 | -0.0348 | No |
| 6 | MYC | MYC Entrez, Source | v-myc myelocytomatosis viral oncogene homolog (avian) | 1609 | 0.108 | -0.0341 | No |
| 7 | KLF10 | KLF10 Entrez, Source | Kruppel-like factor 10 | 1693 | 0.105 | -0.0306 | No |
| 8 | USP15 | USP15 Entrez, Source | ubiquitin specific peptidase 15 | 1732 | 0.104 | -0.0237 | No |
| 9 | TIAM1 | TIAM1 Entrez, Source | T-cell lymphoma invasion and metastasis 1 | 1889 | 0.098 | -0.0264 | No |
| 10 | ACSL3 | ACSL3 Entrez, Source | acyl-CoA synthetase long-chain family member 3 | 1993 | 0.094 | -0.0254 | No |
| 11 | TERF1 | TERF1 Entrez, Source | telomeric repeat binding factor (NIMA-interacting) 1 | 2071 | 0.092 | -0.0226 | No |
| 12 | PRR3 | PRR3 Entrez, Source | proline rich 3 | 2317 | 0.085 | -0.0332 | No |
| 13 | FGF5 | FGF5 Entrez, Source | fibroblast growth factor 5 | 2656 | 0.075 | -0.0518 | No |
| 14 | ZFHX1B | ZFHX1B Entrez, Source | zinc finger homeobox 1b | 2854 | 0.070 | -0.0601 | No |
| 15 | PTPN12 | PTPN12 Entrez, Source | protein tyrosine phosphatase, non-receptor type 12 | 3086 | 0.065 | -0.0715 | No |
| 16 | IGF1R | IGF1R Entrez, Source | insulin-like growth factor 1 receptor | 3241 | 0.061 | -0.0774 | No |
| 17 | FYN | FYN Entrez, Source | FYN oncogene related to SRC, FGR, YES | 3374 | 0.059 | -0.0819 | No |
| 18 | MARK3 | MARK3 Entrez, Source | MAP/microtubule affinity-regulating kinase 3 | 3628 | 0.053 | -0.0960 | No |
| 19 | RBM16 | RBM16 Entrez, Source | RNA binding motif protein 16 | 3735 | 0.051 | -0.0992 | No |
| 20 | ZHX3 | ZHX3 Entrez, Source | zinc fingers and homeoboxes 3 | 3978 | 0.047 | -0.1131 | No |
| 21 | FGF2 | FGF2 Entrez, Source | fibroblast growth factor 2 (basic) | 4030 | 0.046 | -0.1127 | No |
| 22 | EGFR | EGFR Entrez, Source | epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | 4079 | 0.045 | -0.1121 | No |
| 23 | BTRC | BTRC Entrez, Source | beta-transducin repeat containing | 4551 | 0.037 | -0.1442 | No |
| 24 | ATP2B1 | ATP2B1 Entrez, Source | ATPase, Ca++ transporting, plasma membrane 1 | 5439 | 0.023 | -0.2091 | No |
| 25 | MYST3 | MYST3 Entrez, Source | MYST histone acetyltransferase (monocytic leukemia) 3 | 5502 | 0.022 | -0.2117 | No |
| 26 | EIF2C2 | EIF2C2 Entrez, Source | eukaryotic translation initiation factor 2C, 2 | 5725 | 0.018 | -0.2268 | No |
| 27 | CEP350 | CEP350 Entrez, Source | centrosomal protein 350kDa | 5735 | 0.018 | -0.2258 | No |
| 28 | RBM39 | RBM39 Entrez, Source | RNA binding motif protein 39 | 5798 | 0.017 | -0.2288 | No |
| 29 | CENTB2 | CENTB2 Entrez, Source | centaurin, beta 2 | 6012 | 0.014 | -0.2436 | No |
| 30 | RSN | RSN Entrez, Source | restin (Reed-Steinberg cell-expressed intermediate filament-associated protein) | 6102 | 0.013 | -0.2491 | No |
| 31 | TSPAN5 | TSPAN5 Entrez, Source | tetraspanin 5 | 6410 | 0.008 | -0.2716 | No |
| 32 | PHLPP | PHLPP Entrez, Source | PH domain and leucine rich repeat protein phosphatase | 6904 | 0.001 | -0.3087 | No |
| 33 | EDD1 | EDD1 Entrez, Source | E3 ubiquitin protein ligase, HECT domain containing, 1 | 7181 | -0.003 | -0.3293 | No |
| 34 | IL6 | IL6 Entrez, Source | interleukin 6 (interferon, beta 2) | 7239 | -0.004 | -0.3333 | No |
| 35 | ZNF148 | ZNF148 Entrez, Source | zinc finger protein 148 | 7480 | -0.007 | -0.3507 | No |
| 36 | CREB1 | CREB1 Entrez, Source | cAMP responsive element binding protein 1 | 7495 | -0.007 | -0.3511 | No |
| 37 | REV3L | REV3L Entrez, Source | REV3-like, catalytic subunit of DNA polymerase zeta (yeast) | 7625 | -0.009 | -0.3600 | No |
| 38 | AKAP9 | AKAP9 Entrez, Source | A kinase (PRKA) anchor protein (yotiao) 9 | 7704 | -0.010 | -0.3649 | No |
| 39 | CDC2L5 | CDC2L5 Entrez, Source | cell division cycle 2-like 5 (cholinesterase-related cell division controller) | 7727 | -0.011 | -0.3655 | No |
| 40 | TUBGCP3 | TUBGCP3 Entrez, Source | tubulin, gamma complex associated protein 3 | 7754 | -0.011 | -0.3664 | No |
| 41 | ACVR1 | ACVR1 Entrez, Source | activin A receptor, type I | 7793 | -0.012 | -0.3682 | No |
| 42 | MID1 | MID1 Entrez, Source | midline 1 (Opitz/BBB syndrome) | 7962 | -0.014 | -0.3795 | No |
| 43 | RGS7 | RGS7 Entrez, Source | regulator of G-protein signalling 7 | 7991 | -0.015 | -0.3802 | No |
| 44 | GLRB | GLRB Entrez, Source | glycine receptor, beta | 8010 | -0.015 | -0.3802 | No |
| 45 | DYRK1A | DYRK1A Entrez, Source | dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A | 8340 | -0.020 | -0.4031 | No |
| 46 | AFAP | AFAP Entrez, Source | - | 8351 | -0.021 | -0.4019 | No |
| 47 | PARG | PARG Entrez, Source | poly (ADP-ribose) glycohydrolase | 8393 | -0.021 | -0.4030 | No |
| 48 | CREBBP | CREBBP Entrez, Source | CREB binding protein (Rubinstein-Taybi syndrome) | 8561 | -0.024 | -0.4134 | No |
| 49 | NUP153 | NUP153 Entrez, Source | nucleoporin 153kDa | 8693 | -0.026 | -0.4208 | No |
| 50 | PPP1R12A | PPP1R12A Entrez, Source | protein phosphatase 1, regulatory (inhibitor) subunit 12A | 8798 | -0.028 | -0.4261 | No |
| 51 | BMPR1A | BMPR1A Entrez, Source | bone morphogenetic protein receptor, type IA | 8901 | -0.030 | -0.4309 | No |
| 52 | WNT5A | WNT5A Entrez, Source | wingless-type MMTV integration site family, member 5A | 8904 | -0.030 | -0.4283 | No |
| 53 | ATF2 | ATF2 Entrez, Source | activating transcription factor 2 | 9097 | -0.034 | -0.4396 | No |
| 54 | MARCH7 | MARCH7 Entrez, Source | membrane-associated ring finger (C3HC4) 7 | 9196 | -0.036 | -0.4436 | No |
| 55 | PAFAH1B1 | PAFAH1B1 Entrez, Source | platelet-activating factor acetylhydrolase, isoform Ib, alpha subunit 45kDa | 9229 | -0.036 | -0.4427 | No |
| 56 | NCOA3 | NCOA3 Entrez, Source | nuclear receptor coactivator 3 | 9534 | -0.042 | -0.4617 | No |
| 57 | STK3 | STK3 Entrez, Source | serine/threonine kinase 3 (STE20 homolog, yeast) | 9831 | -0.048 | -0.4795 | No |
| 58 | APPBP2 | APPBP2 Entrez, Source | amyloid beta precursor protein (cytoplasmic tail) binding protein 2 | 9900 | -0.050 | -0.4800 | No |
| 59 | PUM2 | PUM2 Entrez, Source | pumilio homolog 2 (Drosophila) | 10034 | -0.053 | -0.4851 | No |
| 60 | E2F5 | E2F5 Entrez, Source | E2F transcription factor 5, p130-binding | 10068 | -0.053 | -0.4826 | No |
| 61 | CNOT4 | CNOT4 Entrez, Source | CCR4-NOT transcription complex, subunit 4 | 10146 | -0.055 | -0.4832 | No |
| 62 | CUTL1 | CUTL1 Entrez, Source | cut-like 1, CCAAT displacement protein (Drosophila) | 10279 | -0.058 | -0.4877 | No |
| 63 | MTMR3 | MTMR3 Entrez, Source | myotubularin related protein 3 | 10836 | -0.073 | -0.5228 | Yes |
| 64 | CNOT2 | CNOT2 Entrez, Source | CCR4-NOT transcription complex, subunit 2 | 10885 | -0.075 | -0.5195 | Yes |
| 65 | SOS2 | SOS2 Entrez, Source | son of sevenless homolog 2 (Drosophila) | 10962 | -0.077 | -0.5180 | Yes |
| 66 | PTPRK | PTPRK Entrez, Source | protein tyrosine phosphatase, receptor type, K | 11105 | -0.082 | -0.5210 | Yes |
| 67 | NFIB | NFIB Entrez, Source | nuclear factor I/B | 11136 | -0.083 | -0.5155 | Yes |
| 68 | BARD1 | BARD1 Entrez, Source | BRCA1 associated RING domain 1 | 11154 | -0.084 | -0.5090 | Yes |
| 69 | STRN3 | STRN3 Entrez, Source | striatin, calmodulin binding protein 3 | 11294 | -0.090 | -0.5111 | Yes |
| 70 | KLF6 | KLF6 Entrez, Source | Kruppel-like factor 6 | 11298 | -0.090 | -0.5028 | Yes |
| 71 | TGFBR3 | TGFBR3 Entrez, Source | transforming growth factor, beta receptor III (betaglycan, 300kDa) | 11551 | -0.102 | -0.5124 | Yes |
| 72 | CLASP2 | CLASP2 Entrez, Source | cytoplasmic linker associated protein 2 | 11749 | -0.111 | -0.5168 | Yes |
| 73 | KIF2A | KIF2A Entrez, Source | kinesin heavy chain member 2A | 11783 | -0.113 | -0.5088 | Yes |
| 74 | ARHGEF7 | ARHGEF7 Entrez, Source | Rho guanine nucleotide exchange factor (GEF) 7 | 11790 | -0.113 | -0.4987 | Yes |
| 75 | IL1RAP | IL1RAP Entrez, Source | interleukin 1 receptor accessory protein | 11842 | -0.116 | -0.4917 | Yes |
| 76 | PTK2 | PTK2 Entrez, Source | PTK2 protein tyrosine kinase 2 | 11904 | -0.119 | -0.4852 | Yes |
| 77 | RASA1 | RASA1 Entrez, Source | RAS p21 protein activator (GTPase activating protein) 1 | 11988 | -0.124 | -0.4799 | Yes |
| 78 | TGFB2 | TGFB2 Entrez, Source | transforming growth factor, beta 2 | 12112 | -0.132 | -0.4769 | Yes |
| 79 | MTF2 | MTF2 Entrez, Source | metal response element binding transcription factor 2 | 12173 | -0.137 | -0.4686 | Yes |
| 80 | CYR61 | CYR61 Entrez, Source | cysteine-rich, angiogenic inducer, 61 | 12269 | -0.146 | -0.4622 | Yes |
| 81 | CTBP2 | CTBP2 Entrez, Source | C-terminal binding protein 2 | 12294 | -0.147 | -0.4502 | Yes |
| 82 | PCTK2 | PCTK2 Entrez, Source | PCTAIRE protein kinase 2 | 12381 | -0.156 | -0.4421 | Yes |
| 83 | GNAQ | GNAQ Entrez, Source | guanine nucleotide binding protein (G protein), q polypeptide | 12403 | -0.158 | -0.4290 | Yes |
| 84 | PLEKHC1 | PLEKHC1 Entrez, Source | pleckstrin homology domain containing, family C (with FERM domain) member 1 | 12574 | -0.181 | -0.4249 | Yes |
| 85 | TRIM2 | TRIM2 Entrez, Source | tripartite motif-containing 2 | 12579 | -0.181 | -0.4083 | Yes |
| 86 | CBLB | CBLB Entrez, Source | Cas-Br-M (murine) ecotropic retroviral transforming sequence b | 12594 | -0.184 | -0.3922 | Yes |
| 87 | VEGFC | VEGFC Entrez, Source | vascular endothelial growth factor C | 12697 | -0.204 | -0.3808 | Yes |
| 88 | NIPBL | NIPBL Entrez, Source | Nipped-B homolog (Drosophila) | 12794 | -0.223 | -0.3673 | Yes |
| 89 | TLE4 | TLE4 Entrez, Source | transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) | 12889 | -0.247 | -0.3513 | Yes |
| 90 | CEP170 | CEP170 Entrez, Source | centrosomal protein 170kDa | 12890 | -0.249 | -0.3280 | Yes |
| 91 | UGCG | UGCG Entrez, Source | UDP-glucose ceramide glucosyltransferase | 12906 | -0.254 | -0.3054 | Yes |
| 92 | TRIO | TRIO Entrez, Source | triple functional domain (PTPRF interacting) | 12955 | -0.274 | -0.2834 | Yes |
| 93 | ZNF292 | ZNF292 Entrez, Source | zinc finger protein 292 | 13006 | -0.293 | -0.2598 | Yes |
| 94 | RRAS2 | RRAS2 Entrez, Source | related RAS viral (r-ras) oncogene homolog 2 | 13055 | -0.322 | -0.2333 | Yes |
| 95 | RUNX1 | RUNX1 Entrez, Source | runt-related transcription factor 1 (acute myeloid leukemia 1; aml1 oncogene) | 13073 | -0.336 | -0.2033 | Yes |
| 96 | SCHIP1 | SCHIP1 Entrez, Source | schwannomin interacting protein 1 | 13180 | -0.438 | -0.1703 | Yes |
| 97 | CTGF | CTGF Entrez, Source | connective tissue growth factor | 13260 | -0.571 | -0.1229 | Yes |
| 98 | GNAI1 | GNAI1 Entrez, Source | guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 | 13262 | -0.580 | -0.0687 | Yes |
| 99 | F3 | F3 Entrez, Source | coagulation factor III (thromboplastin, tissue factor) | 13308 | -0.800 | 0.0026 | Yes |