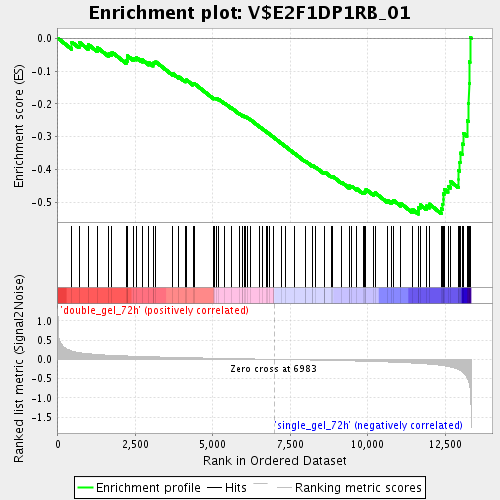
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_72h_versus_single_gel_72h.class.cls #double_gel_72h_versus_single_gel_72h_repos |
| Phenotype | class.cls#double_gel_72h_versus_single_gel_72h_repos |
| Upregulated in class | single_gel_72h |
| GeneSet | V$E2F1DP1RB_01 |
| Enrichment Score (ES) | -0.5366827 |
| Normalized Enrichment Score (NES) | -1.8324943 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.031302754 |
| FWER p-Value | 0.877 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | ALDH6A1 | ALDH6A1 Entrez, Source | aldehyde dehydrogenase 6 family, member A1 | 445 | 0.217 | -0.0115 | No |
| 2 | DMD | DMD Entrez, Source | dystrophin (muscular dystrophy, Duchenne and Becker types) | 686 | 0.175 | -0.0119 | No |
| 3 | ATF5 | ATF5 Entrez, Source | activating transcription factor 5 | 979 | 0.147 | -0.0190 | No |
| 4 | NOLC1 | NOLC1 Entrez, Source | nucleolar and coiled-body phosphoprotein 1 | 1277 | 0.125 | -0.0287 | No |
| 5 | PCSK1 | PCSK1 Entrez, Source | proprotein convertase subtilisin/kexin type 1 | 1646 | 0.107 | -0.0457 | No |
| 6 | UGCGL1 | UGCGL1 Entrez, Source | UDP-glucose ceramide glucosyltransferase-like 1 | 1741 | 0.104 | -0.0423 | No |
| 7 | SFRS7 | SFRS7 Entrez, Source | splicing factor, arginine/serine-rich 7, 35kDa | 2205 | 0.088 | -0.0683 | No |
| 8 | GRIA4 | GRIA4 Entrez, Source | glutamate receptor, ionotrophic, AMPA 4 | 2238 | 0.087 | -0.0619 | No |
| 9 | SLC6A4 | SLC6A4 Entrez, Source | solute carrier family 6 (neurotransmitter transporter, serotonin), member 4 | 2243 | 0.086 | -0.0535 | No |
| 10 | AK2 | AK2 Entrez, Source | adenylate kinase 2 | 2453 | 0.081 | -0.0610 | No |
| 11 | CDC25A | CDC25A Entrez, Source | cell division cycle 25A | 2527 | 0.079 | -0.0585 | No |
| 12 | SLC38A1 | SLC38A1 Entrez, Source | solute carrier family 38, member 1 | 2719 | 0.073 | -0.0655 | No |
| 13 | HMGN2 | HMGN2 Entrez, Source | high-mobility group nucleosomal binding domain 2 | 2936 | 0.068 | -0.0749 | No |
| 14 | SLC9A5 | SLC9A5 Entrez, Source | solute carrier family 9 (sodium/hydrogen exchanger), member 5 | 3069 | 0.065 | -0.0783 | No |
| 15 | FMO4 | FMO4 Entrez, Source | flavin containing monooxygenase 4 | 3098 | 0.064 | -0.0739 | No |
| 16 | UNG | UNG Entrez, Source | uracil-DNA glycosylase | 3161 | 0.063 | -0.0722 | No |
| 17 | MXD3 | MXD3 Entrez, Source | MAX dimerization protein 3 | 3701 | 0.052 | -0.1076 | No |
| 18 | IMPDH2 | IMPDH2 Entrez, Source | IMP (inosine monophosphate) dehydrogenase 2 | 3898 | 0.048 | -0.1175 | No |
| 19 | POU4F1 | POU4F1 Entrez, Source | POU domain, class 4, transcription factor 1 | 4112 | 0.044 | -0.1291 | No |
| 20 | XTP3TPA | XTP3TPA Entrez, Source | - | 4144 | 0.044 | -0.1269 | No |
| 21 | PAX6 | PAX6 Entrez, Source | paired box gene 6 (aniridia, keratitis) | 4359 | 0.040 | -0.1390 | No |
| 22 | CORT | CORT Entrez, Source | cortistatin | 4395 | 0.040 | -0.1377 | No |
| 23 | ATRX | ATRX Entrez, Source | alpha thalassemia/mental retardation syndrome X-linked (RAD54 homolog, S. cerevisiae) | 5030 | 0.029 | -0.1826 | No |
| 24 | ACBD6 | ACBD6 Entrez, Source | acyl-Coenzyme A binding domain containing 6 | 5066 | 0.028 | -0.1824 | No |
| 25 | HCN3 | HCN3 Entrez, Source | hyperpolarization activated cyclic nucleotide-gated potassium channel 3 | 5113 | 0.027 | -0.1831 | No |
| 26 | HOXC10 | HOXC10 Entrez, Source | homeobox C10 | 5188 | 0.026 | -0.1860 | No |
| 27 | NPR3 | NPR3 Entrez, Source | natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) | 5375 | 0.024 | -0.1977 | No |
| 28 | NASP | NASP Entrez, Source | nuclear autoantigenic sperm protein (histone-binding) | 5597 | 0.020 | -0.2123 | No |
| 29 | MAPK6 | MAPK6 Entrez, Source | mitogen-activated protein kinase 6 | 5856 | 0.016 | -0.2301 | No |
| 30 | PCNA | PCNA Entrez, Source | proliferating cell nuclear antigen | 5968 | 0.015 | -0.2370 | No |
| 31 | SFRS2 | SFRS2 Entrez, Source | splicing factor, arginine/serine-rich 2 | 5972 | 0.015 | -0.2357 | No |
| 32 | DPYSL2 | DPYSL2 Entrez, Source | dihydropyrimidinase-like 2 | 6036 | 0.014 | -0.2391 | No |
| 33 | HNRPA1 | HNRPA1 Entrez, Source | heterogeneous nuclear ribonucleoprotein A1 | 6063 | 0.013 | -0.2397 | No |
| 34 | CTDSP1 | CTDSP1 Entrez, Source | CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 1 | 6119 | 0.013 | -0.2426 | No |
| 35 | PIM1 | PIM1 Entrez, Source | pim-1 oncogene | 6208 | 0.011 | -0.2481 | No |
| 36 | SP3 | SP3 Entrez, Source | Sp3 transcription factor | 6515 | 0.006 | -0.2706 | No |
| 37 | SUMO1 | SUMO1 Entrez, Source | SMT3 suppressor of mif two 3 homolog 1 (S. cerevisiae) | 6611 | 0.005 | -0.2772 | No |
| 38 | GSPT1 | GSPT1 Entrez, Source | G1 to S phase transition 1 | 6717 | 0.004 | -0.2848 | No |
| 39 | SEZ6 | SEZ6 Entrez, Source | seizure related 6 homolog (mouse) | 6779 | 0.003 | -0.2891 | No |
| 40 | PRPS1 | PRPS1 Entrez, Source | phosphoribosyl pyrophosphate synthetase 1 | 6821 | 0.002 | -0.2920 | No |
| 41 | NCL | NCL Entrez, Source | nucleolin | 6959 | 0.000 | -0.3023 | No |
| 42 | DCK | DCK Entrez, Source | deoxycytidine kinase | 6967 | 0.000 | -0.3028 | No |
| 43 | USP52 | USP52 Entrez, Source | ubiquitin specific peptidase 52 | 7214 | -0.003 | -0.3210 | No |
| 44 | POLD3 | POLD3 Entrez, Source | polymerase (DNA-directed), delta 3, accessory subunit | 7338 | -0.005 | -0.3298 | No |
| 45 | CTCF | CTCF Entrez, Source | CCCTC-binding factor (zinc finger protein) | 7648 | -0.010 | -0.3521 | No |
| 46 | PPIG | PPIG Entrez, Source | peptidylprolyl isomerase G (cyclophilin G) | 7975 | -0.015 | -0.3752 | No |
| 47 | NRP2 | NRP2 Entrez, Source | neuropilin 2 | 8004 | -0.015 | -0.3758 | No |
| 48 | NELL2 | NELL2 Entrez, Source | NEL-like 2 (chicken) | 8206 | -0.018 | -0.3892 | No |
| 49 | PPP1CC | PPP1CC Entrez, Source | protein phosphatase 1, catalytic subunit, gamma isoform | 8221 | -0.018 | -0.3884 | No |
| 50 | EIF4A1 | EIF4A1 Entrez, Source | eukaryotic translation initiation factor 4A, isoform 1 | 8320 | -0.020 | -0.3937 | No |
| 51 | TLE3 | TLE3 Entrez, Source | transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila) | 8587 | -0.025 | -0.4113 | No |
| 52 | EPHB1 | EPHB1 Entrez, Source | EPH receptor B1 | 8589 | -0.025 | -0.4089 | No |
| 53 | RET | RET Entrez, Source | ret proto-oncogene (multiple endocrine neoplasia and medullary thyroid carcinoma 1, Hirschsprung disease) | 8617 | -0.025 | -0.4084 | No |
| 54 | KCNA6 | KCNA6 Entrez, Source | potassium voltage-gated channel, shaker-related subfamily, member 6 | 8822 | -0.029 | -0.4209 | No |
| 55 | KBTBD7 | KBTBD7 Entrez, Source | kelch repeat and BTB (POZ) domain containing 7 | 8874 | -0.029 | -0.4218 | No |
| 56 | GABRB3 | GABRB3 Entrez, Source | gamma-aminobutyric acid (GABA) A receptor, beta 3 | 9166 | -0.035 | -0.4402 | No |
| 57 | DNMT1 | DNMT1 Entrez, Source | DNA (cytosine-5-)-methyltransferase 1 | 9405 | -0.040 | -0.4541 | No |
| 58 | CLTA | CLTA Entrez, Source | clathrin, light chain (Lca) | 9412 | -0.040 | -0.4505 | No |
| 59 | LHX5 | LHX5 Entrez, Source | LIM homeobox 5 | 9484 | -0.041 | -0.4517 | No |
| 60 | RFC1 | RFC1 Entrez, Source | replication factor C (activator 1) 1, 145kDa | 9644 | -0.044 | -0.4592 | No |
| 61 | PVRL1 | PVRL1 Entrez, Source | poliovirus receptor-related 1 (herpesvirus entry mediator C; nectin) | 9846 | -0.049 | -0.4694 | No |
| 62 | POLD1 | POLD1 Entrez, Source | polymerase (DNA directed), delta 1, catalytic subunit 125kDa | 9909 | -0.050 | -0.4690 | No |
| 63 | KPNB1 | KPNB1 Entrez, Source | karyopherin (importin) beta 1 | 9930 | -0.051 | -0.4653 | No |
| 64 | KLF4 | KLF4 Entrez, Source | Kruppel-like factor 4 (gut) | 9940 | -0.051 | -0.4609 | No |
| 65 | ILF3 | ILF3 Entrez, Source | interleukin enhancer binding factor 3, 90kDa | 10199 | -0.057 | -0.4746 | No |
| 66 | HNRPR | HNRPR Entrez, Source | heterogeneous nuclear ribonucleoprotein R | 10233 | -0.058 | -0.4712 | No |
| 67 | RAB11B | RAB11B Entrez, Source | RAB11B, member RAS oncogene family | 10624 | -0.068 | -0.4938 | No |
| 68 | ING3 | ING3 Entrez, Source | inhibitor of growth family, member 3 | 10772 | -0.072 | -0.4977 | No |
| 69 | MAPT | MAPT Entrez, Source | microtubule-associated protein tau | 10830 | -0.073 | -0.4945 | No |
| 70 | H2AFZ | H2AFZ Entrez, Source | H2A histone family, member Z | 11071 | -0.081 | -0.5045 | No |
| 71 | RBBP7 | RBBP7 Entrez, Source | retinoblastoma binding protein 7 | 11453 | -0.098 | -0.5233 | No |
| 72 | LIG1 | LIG1 Entrez, Source | ligase I, DNA, ATP-dependent | 11631 | -0.106 | -0.5260 | Yes |
| 73 | RHD | RHD Entrez, Source | Rh blood group, D antigen | 11643 | -0.106 | -0.5160 | Yes |
| 74 | CDCA7 | CDCA7 Entrez, Source | cell division cycle associated 7 | 11689 | -0.109 | -0.5084 | Yes |
| 75 | SMAD6 | SMAD6 Entrez, Source | SMAD, mothers against DPP homolog 6 (Drosophila) | 11893 | -0.118 | -0.5117 | Yes |
| 76 | NUP62 | NUP62 Entrez, Source | nucleoporin 62kDa | 11981 | -0.124 | -0.5058 | Yes |
| 77 | E2F1 | E2F1 Entrez, Source | E2F transcription factor 1 | 12376 | -0.156 | -0.5197 | Yes |
| 78 | MCM7 | MCM7 Entrez, Source | MCM7 minichromosome maintenance deficient 7 (S. cerevisiae) | 12413 | -0.160 | -0.5063 | Yes |
| 79 | MAT2A | MAT2A Entrez, Source | methionine adenosyltransferase II, alpha | 12429 | -0.161 | -0.4910 | Yes |
| 80 | MSH2 | MSH2 Entrez, Source | mutS homolog 2, colon cancer, nonpolyposis type 1 (E. coli) | 12432 | -0.162 | -0.4748 | Yes |
| 81 | PAQR4 | PAQR4 Entrez, Source | progestin and adipoQ receptor family member IV | 12466 | -0.166 | -0.4605 | Yes |
| 82 | TMPO | TMPO Entrez, Source | thymopoietin | 12590 | -0.183 | -0.4512 | Yes |
| 83 | POLA2 | POLA2 Entrez, Source | polymerase (DNA directed), alpha 2 (70kD subunit) | 12673 | -0.198 | -0.4373 | Yes |
| 84 | WEE1 | WEE1 Entrez, Source | WEE1 homolog (S. pombe) | 12929 | -0.262 | -0.4300 | Yes |
| 85 | SLCO3A1 | SLCO3A1 Entrez, Source | solute carrier organic anion transporter family, member 3A1 | 12932 | -0.263 | -0.4035 | Yes |
| 86 | MYH10 | MYH10 Entrez, Source | myosin, heavy chain 10, non-muscle | 12969 | -0.279 | -0.3779 | Yes |
| 87 | TYRO3 | TYRO3 Entrez, Source | TYRO3 protein tyrosine kinase | 12984 | -0.284 | -0.3501 | Yes |
| 88 | HNRPD | HNRPD Entrez, Source | heterogeneous nuclear ribonucleoprotein D (AU-rich element RNA binding protein 1, 37kDa) | 13048 | -0.319 | -0.3226 | Yes |
| 89 | ONECUT1 | ONECUT1 Entrez, Source | one cut domain, family member 1 | 13087 | -0.346 | -0.2903 | Yes |
| 90 | ID3 | ID3 Entrez, Source | inhibitor of DNA binding 3, dominant negative helix-loop-helix protein | 13204 | -0.471 | -0.2513 | Yes |
| 91 | SOAT1 | SOAT1 Entrez, Source | sterol O-acyltransferase (acyl-Coenzyme A: cholesterol acyltransferase) 1 | 13248 | -0.551 | -0.1988 | Yes |
| 92 | MCM6 | MCM6 Entrez, Source | MCM6 minichromosome maintenance deficient 6 (MIS5 homolog, S. pombe) (S. cerevisiae) | 13267 | -0.614 | -0.1379 | Yes |
| 93 | KIAA0101 | KIAA0101 Entrez, Source | KIAA0101 | 13280 | -0.659 | -0.0719 | Yes |
| 94 | STMN1 | STMN1 Entrez, Source | stathmin 1/oncoprotein 18 | 13302 | -0.755 | 0.0030 | Yes |

