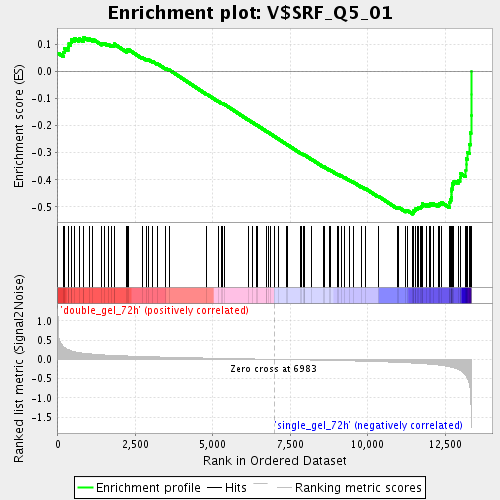
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_72h_versus_single_gel_72h.class.cls #double_gel_72h_versus_single_gel_72h_repos |
| Phenotype | class.cls#double_gel_72h_versus_single_gel_72h_repos |
| Upregulated in class | single_gel_72h |
| GeneSet | V$SRF_Q5_01 |
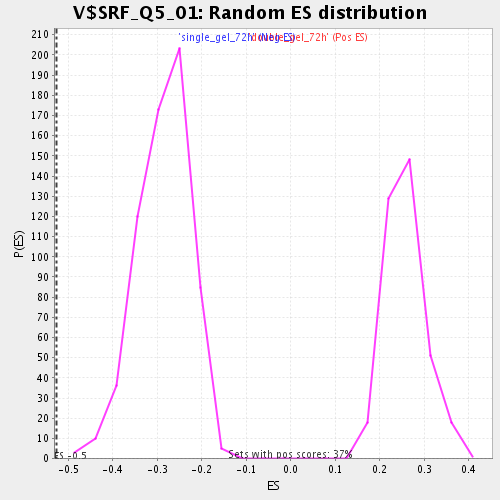
| Enrichment Score (ES) | -0.5265778 |
| Normalized Enrichment Score (NES) | -1.8475229 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.028190237 |
| FWER p-Value | 0.824 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | FOS | FOS Entrez, Source | v-fos FBJ murine osteosarcoma viral oncogene homolog | 0 | 1.284 | 0.0682 | No |
| 2 | EGR2 | EGR2 Entrez, Source | early growth response 2 (Krox-20 homolog, Drosophila) | 180 | 0.328 | 0.0721 | No |
| 3 | RARB | RARB Entrez, Source | retinoic acid receptor, beta | 214 | 0.304 | 0.0858 | No |
| 4 | PPAP2B | PPAP2B Entrez, Source | phosphatidic acid phosphatase type 2B | 329 | 0.257 | 0.0908 | No |
| 5 | ABCA1 | ABCA1 Entrez, Source | ATP-binding cassette, sub-family A (ABC1), member 1 | 355 | 0.248 | 0.1021 | No |
| 6 | SLC16A6 | SLC16A6 Entrez, Source | solute carrier family 16, member 6 (monocarboxylic acid transporter 7) | 422 | 0.224 | 0.1090 | No |
| 7 | EGR1 | EGR1 Entrez, Source | early growth response 1 | 437 | 0.219 | 0.1196 | No |
| 8 | FGF17 | FGF17 Entrez, Source | fibroblast growth factor 17 | 538 | 0.195 | 0.1224 | No |
| 9 | SRD5A2 | SRD5A2 Entrez, Source | steroid-5-alpha-reductase, alpha polypeptide 2 (3-oxo-5 alpha-steroid delta 4-dehydrogenase alpha 2) | 681 | 0.176 | 0.1210 | No |
| 10 | GADD45G | GADD45G Entrez, Source | growth arrest and DNA-damage-inducible, gamma | 829 | 0.160 | 0.1184 | No |
| 11 | IER2 | IER2 Entrez, Source | immediate early response 2 | 833 | 0.159 | 0.1266 | No |
| 12 | FLNC | FLNC Entrez, Source | filamin C, gamma (actin binding protein 280) | 1014 | 0.144 | 0.1207 | No |
| 13 | FGF7 | FGF7 Entrez, Source | fibroblast growth factor 7 (keratinocyte growth factor) | 1129 | 0.135 | 0.1192 | No |
| 14 | ATF3 | ATF3 Entrez, Source | activating transcription factor 3 | 1413 | 0.118 | 0.1041 | No |
| 15 | JUNB | JUNB Entrez, Source | jun B proto-oncogene | 1487 | 0.114 | 0.1046 | No |
| 16 | COL1A1 | COL1A1 Entrez, Source | collagen, type I, alpha 1 | 1619 | 0.108 | 0.1005 | No |
| 17 | ASPA | ASPA Entrez, Source | aspartoacylase (Canavan disease) | 1733 | 0.104 | 0.0974 | No |
| 18 | OTX1 | OTX1 Entrez, Source | orthodenticle homolog 1 (Drosophila) | 1813 | 0.101 | 0.0968 | No |
| 19 | MAPK14 | MAPK14 Entrez, Source | mitogen-activated protein kinase 14 | 1814 | 0.101 | 0.1022 | No |
| 20 | DUSP5 | DUSP5 Entrez, Source | dual specificity phosphatase 5 | 2225 | 0.087 | 0.0758 | No |
| 21 | FOXE3 | FOXE3 Entrez, Source | forkhead box E3 | 2231 | 0.087 | 0.0801 | No |
| 22 | MBNL1 | MBNL1 Entrez, Source | muscleblind-like (Drosophila) | 2291 | 0.085 | 0.0802 | No |
| 23 | TNMD | TNMD Entrez, Source | tenomodulin | 2726 | 0.073 | 0.0513 | No |
| 24 | COL23A1 | COL23A1 Entrez, Source | collagen, type XXIII, alpha 1 | 2873 | 0.070 | 0.0439 | No |
| 25 | CSDA | CSDA Entrez, Source | cold shock domain protein A | 2927 | 0.068 | 0.0435 | No |
| 26 | ATF4 | ATF4 Entrez, Source | activating transcription factor 4 (tax-responsive enhancer element B67) | 3058 | 0.065 | 0.0372 | No |
| 27 | NPPA | NPPA Entrez, Source | natriuretic peptide precursor A | 3206 | 0.062 | 0.0294 | No |
| 28 | FGF8 | FGF8 Entrez, Source | fibroblast growth factor 8 (androgen-induced) | 3478 | 0.056 | 0.0119 | No |
| 29 | PPARG | PPARG Entrez, Source | peroxisome proliferative activated receptor, gamma | 3592 | 0.054 | 0.0062 | No |
| 30 | STAT5B | STAT5B Entrez, Source | signal transducer and activator of transcription 5B | 4806 | 0.032 | -0.0837 | No |
| 31 | NRXN3 | NRXN3 Entrez, Source | neurexin 3 | 5181 | 0.027 | -0.1106 | No |
| 32 | PADI2 | PADI2 Entrez, Source | peptidyl arginine deiminase, type II | 5275 | 0.025 | -0.1163 | No |
| 33 | ARF6 | ARF6 Entrez, Source | ADP-ribosylation factor 6 | 5321 | 0.024 | -0.1184 | No |
| 34 | NPR3 | NPR3 Entrez, Source | natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) | 5375 | 0.024 | -0.1211 | No |
| 35 | TRDN | TRDN Entrez, Source | triadin | 6145 | 0.012 | -0.1786 | No |
| 36 | FGFRL1 | FGFRL1 Entrez, Source | fibroblast growth factor receptor-like 1 | 6270 | 0.010 | -0.1874 | No |
| 37 | GGN | GGN Entrez, Source | gametogenetin | 6394 | 0.008 | -0.1963 | No |
| 38 | PLCB3 | PLCB3 Entrez, Source | phospholipase C, beta 3 (phosphatidylinositol-specific) | 6441 | 0.007 | -0.1994 | No |
| 39 | MYH11 | MYH11 Entrez, Source | myosin, heavy chain 11, smooth muscle | 6721 | 0.004 | -0.2203 | No |
| 40 | KCNK3 | KCNK3 Entrez, Source | potassium channel, subfamily K, member 3 | 6725 | 0.004 | -0.2203 | No |
| 41 | EGR3 | EGR3 Entrez, Source | early growth response 3 | 6801 | 0.003 | -0.2259 | No |
| 42 | CACNA1B | CACNA1B Entrez, Source | calcium channel, voltage-dependent, L type, alpha 1B subunit | 6851 | 0.002 | -0.2294 | No |
| 43 | RHOJ | RHOJ Entrez, Source | ras homolog gene family, member J | 7002 | -0.000 | -0.2408 | No |
| 44 | MAP1A | MAP1A Entrez, Source | microtubule-associated protein 1A | 7116 | -0.002 | -0.2492 | No |
| 45 | NR2F2 | NR2F2 Entrez, Source | nuclear receptor subfamily 2, group F, member 2 | 7382 | -0.006 | -0.2689 | No |
| 46 | IL17B | IL17B Entrez, Source | interleukin 17B | 7417 | -0.006 | -0.2712 | No |
| 47 | DLL1 | DLL1 Entrez, Source | delta-like 1 (Drosophila) | 7827 | -0.013 | -0.3014 | No |
| 48 | NFATC4 | NFATC4 Entrez, Source | nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 | 7871 | -0.013 | -0.3040 | No |
| 49 | DGKG | DGKG Entrez, Source | diacylglycerol kinase, gamma 90kDa | 7914 | -0.014 | -0.3064 | No |
| 50 | PRKAB2 | PRKAB2 Entrez, Source | protein kinase, AMP-activated, beta 2 non-catalytic subunit | 7945 | -0.014 | -0.3079 | No |
| 51 | MID1 | MID1 Entrez, Source | midline 1 (Opitz/BBB syndrome) | 7962 | -0.014 | -0.3083 | No |
| 52 | NR2F1 | NR2F1 Entrez, Source | nuclear receptor subfamily 2, group F, member 1 | 8175 | -0.018 | -0.3234 | No |
| 53 | CKM | CKM Entrez, Source | creatine kinase, muscle | 8557 | -0.024 | -0.3509 | No |
| 54 | TLE3 | TLE3 Entrez, Source | transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila) | 8587 | -0.025 | -0.3518 | No |
| 55 | MRGPRF | MRGPRF Entrez, Source | MAS-related GPR, member F | 8754 | -0.027 | -0.3629 | No |
| 56 | PPP1R12A | PPP1R12A Entrez, Source | protein phosphatase 1, regulatory (inhibitor) subunit 12A | 8798 | -0.028 | -0.3647 | No |
| 57 | GPR20 | GPR20 Entrez, Source | G protein-coupled receptor 20 | 9010 | -0.032 | -0.3789 | No |
| 58 | ANXA6 | ANXA6 Entrez, Source | annexin A6 | 9062 | -0.033 | -0.3810 | No |
| 59 | CRH | CRH Entrez, Source | corticotropin releasing hormone | 9147 | -0.035 | -0.3855 | No |
| 60 | MAP2K6 | MAP2K6 Entrez, Source | mitogen-activated protein kinase kinase 6 | 9252 | -0.037 | -0.3914 | No |
| 61 | SLC4A3 | SLC4A3 Entrez, Source | solute carrier family 4, anion exchanger, member 3 | 9406 | -0.040 | -0.4008 | No |
| 62 | PHF12 | PHF12 Entrez, Source | PHD finger protein 12 | 9531 | -0.042 | -0.4080 | No |
| 63 | SLC2A4 | SLC2A4 Entrez, Source | solute carrier family 2 (facilitated glucose transporter), member 4 | 9794 | -0.048 | -0.4252 | No |
| 64 | TAF5L | TAF5L Entrez, Source | TAF5-like RNA polymerase II, p300/CBP-associated factor (PCAF)-associated factor, 65kDa | 9912 | -0.050 | -0.4314 | No |
| 65 | KCNA1 | KCNA1 Entrez, Source | potassium voltage-gated channel, shaker-related subfamily, member 1 (episodic ataxia with myokymia) | 10336 | -0.060 | -0.4602 | No |
| 66 | RIMS1 | RIMS1 Entrez, Source | regulating synaptic membrane exocytosis 1 | 10964 | -0.077 | -0.5034 | No |
| 67 | KCNH2 | KCNH2 Entrez, Source | potassium voltage-gated channel, subfamily H (eag-related), member 2 | 10984 | -0.078 | -0.5007 | No |
| 68 | CSRP1 | CSRP1 Entrez, Source | cysteine and glycine-rich protein 1 | 11225 | -0.087 | -0.5142 | No |
| 69 | MUS81 | MUS81 Entrez, Source | MUS81 endonuclease homolog (S. cerevisiae) | 11274 | -0.089 | -0.5131 | No |
| 70 | RBBP7 | RBBP7 Entrez, Source | retinoblastoma binding protein 7 | 11453 | -0.098 | -0.5214 | Yes |
| 71 | ACTG2 | ACTG2 Entrez, Source | actin, gamma 2, smooth muscle, enteric | 11475 | -0.099 | -0.5178 | Yes |
| 72 | RERE | RERE Entrez, Source | arginine-glutamic acid dipeptide (RE) repeats | 11485 | -0.099 | -0.5132 | Yes |
| 73 | TRIM3 | TRIM3 Entrez, Source | tripartite motif-containing 3 | 11524 | -0.100 | -0.5107 | Yes |
| 74 | ACTB | ACTB Entrez, Source | actin, beta | 11552 | -0.102 | -0.5073 | Yes |
| 75 | SDPR | SDPR Entrez, Source | serum deprivation response (phosphatidylserine binding protein) | 11603 | -0.105 | -0.5056 | Yes |
| 76 | DIXDC1 | DIXDC1 Entrez, Source | DIX domain containing 1 | 11625 | -0.106 | -0.5015 | Yes |
| 77 | ITGB6 | ITGB6 Entrez, Source | integrin, beta 6 | 11708 | -0.110 | -0.5019 | Yes |
| 78 | MAPKAPK2 | MAPKAPK2 Entrez, Source | mitogen-activated protein kinase-activated protein kinase 2 | 11727 | -0.110 | -0.4974 | Yes |
| 79 | PFN1 | PFN1 Entrez, Source | profilin 1 | 11762 | -0.112 | -0.4940 | Yes |
| 80 | TGFB1I1 | TGFB1I1 Entrez, Source | transforming growth factor beta 1 induced transcript 1 | 11764 | -0.112 | -0.4881 | Yes |
| 81 | CDKN1B | CDKN1B Entrez, Source | cyclin-dependent kinase inhibitor 1B (p27, Kip1) | 11886 | -0.118 | -0.4910 | Yes |
| 82 | TPM3 | TPM3 Entrez, Source | tropomyosin 3 | 11979 | -0.123 | -0.4914 | Yes |
| 83 | ACTR3 | ACTR3 Entrez, Source | ARP3 actin-related protein 3 homolog (yeast) | 12015 | -0.126 | -0.4874 | Yes |
| 84 | NRAS | NRAS Entrez, Source | neuroblastoma RAS viral (v-ras) oncogene homolog | 12113 | -0.132 | -0.4877 | Yes |
| 85 | GRK6 | GRK6 Entrez, Source | G protein-coupled receptor kinase 6 | 12273 | -0.146 | -0.4919 | Yes |
| 86 | H2AFX | H2AFX Entrez, Source | H2A histone family, member X | 12313 | -0.149 | -0.4869 | Yes |
| 87 | CDC25B | CDC25B Entrez, Source | cell division cycle 25B | 12377 | -0.156 | -0.4834 | Yes |
| 88 | PICALM | PICALM Entrez, Source | phosphatidylinositol binding clathrin assembly protein | 12640 | -0.193 | -0.4930 | Yes |
| 89 | ASB2 | ASB2 Entrez, Source | ankyrin repeat and SOCS box-containing 2 | 12641 | -0.193 | -0.4828 | Yes |
| 90 | FOXP1 | FOXP1 Entrez, Source | forkhead box P1 | 12657 | -0.196 | -0.4735 | Yes |
| 91 | CALD1 | CALD1 Entrez, Source | caldesmon 1 | 12690 | -0.203 | -0.4651 | Yes |
| 92 | PLN | PLN Entrez, Source | phospholamban | 12709 | -0.206 | -0.4555 | Yes |
| 93 | SLC25A4 | SLC25A4 Entrez, Source | solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 4 | 12710 | -0.207 | -0.4445 | Yes |
| 94 | DSTN | DSTN Entrez, Source | destrin (actin depolymerizing factor) | 12715 | -0.208 | -0.4338 | Yes |
| 95 | MYO1E | MYO1E Entrez, Source | myosin IE | 12726 | -0.209 | -0.4234 | Yes |
| 96 | SGK | SGK Entrez, Source | serum/glucocorticoid regulated kinase | 12735 | -0.210 | -0.4129 | Yes |
| 97 | CFL1 | CFL1 Entrez, Source | cofilin 1 (non-muscle) | 12779 | -0.218 | -0.4045 | Yes |
| 98 | MYADM | MYADM Entrez, Source | myeloid-associated differentiation marker | 12919 | -0.258 | -0.4013 | Yes |
| 99 | TIMP3 | TIMP3 Entrez, Source | TIMP metallopeptidase inhibitor 3 (Sorsby fundus dystrophy, pseudoinflammatory) | 12978 | -0.283 | -0.3906 | Yes |
| 100 | CXCL5 | CXCL5 Entrez, Source | chemokine (C-X-C motif) ligand 5 | 12997 | -0.289 | -0.3767 | Yes |
| 101 | LCP1 | LCP1 Entrez, Source | lymphocyte cytosolic protein 1 (L-plastin) | 13166 | -0.425 | -0.3668 | Yes |
| 102 | CNN1 | CNN1 Entrez, Source | calponin 1, basic, smooth muscle | 13171 | -0.429 | -0.3443 | Yes |
| 103 | PDLIM7 | PDLIM7 Entrez, Source | PDZ and LIM domain 7 (enigma) | 13173 | -0.431 | -0.3215 | Yes |
| 104 | CITED2 | CITED2 Entrez, Source | Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 | 13211 | -0.481 | -0.2987 | Yes |
| 105 | ACTN1 | ACTN1 Entrez, Source | actinin, alpha 1 | 13279 | -0.655 | -0.2690 | Yes |
| 106 | PPP2R3A | PPP2R3A Entrez, Source | protein phosphatase 2 (formerly 2A), regulatory subunit B'', alpha | 13312 | -0.847 | -0.2264 | Yes |
| 107 | CA3 | CA3 Entrez, Source | carbonic anhydrase III, muscle specific | 13334 | -1.240 | -0.1621 | Yes |
| 108 | TAGLN | TAGLN Entrez, Source | transgelin | 13339 | -1.475 | -0.0841 | Yes |
| 109 | ACTA1 | ACTA1 Entrez, Source | actin, alpha 1, skeletal muscle | 13340 | -1.586 | 0.0002 | Yes |