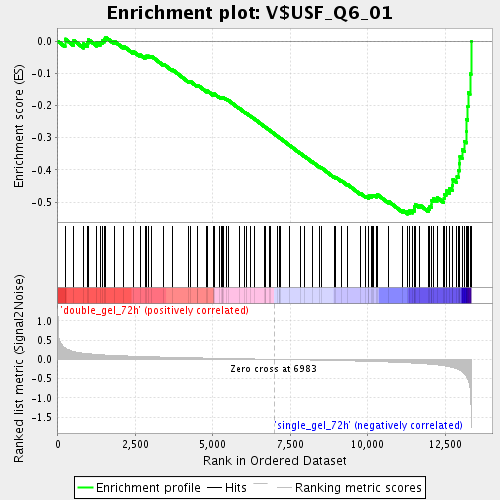
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_72h_versus_single_gel_72h.class.cls #double_gel_72h_versus_single_gel_72h_repos |
| Phenotype | class.cls#double_gel_72h_versus_single_gel_72h_repos |
| Upregulated in class | single_gel_72h |
| GeneSet | V$USF_Q6_01 |
| Enrichment Score (ES) | -0.5375257 |
| Normalized Enrichment Score (NES) | -1.8692642 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.027252456 |
| FWER p-Value | 0.734 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | ACACA | ACACA Entrez, Source | acetyl-Coenzyme A carboxylase alpha | 231 | 0.293 | 0.0070 | No |
| 2 | SC5DL | SC5DL Entrez, Source | sterol-C5-desaturase (ERG3 delta-5-desaturase homolog, fungal)-like | 512 | 0.201 | 0.0026 | No |
| 3 | GADD45G | GADD45G Entrez, Source | growth arrest and DNA-damage-inducible, gamma | 829 | 0.160 | -0.0080 | No |
| 4 | HMOX1 | HMOX1 Entrez, Source | heme oxygenase (decycling) 1 | 949 | 0.149 | -0.0045 | No |
| 5 | SLC16A1 | SLC16A1 Entrez, Source | solute carrier family 16, member 1 (monocarboxylic acid transporter 1) | 974 | 0.147 | 0.0060 | No |
| 6 | HTF9C | HTF9C Entrez, Source | - | 1256 | 0.127 | -0.0047 | No |
| 7 | PDP2 | PDP2 Entrez, Source | - | 1362 | 0.121 | -0.0025 | No |
| 8 | SSR1 | SSR1 Entrez, Source | signal sequence receptor, alpha (translocon-associated protein alpha) | 1437 | 0.116 | 0.0016 | No |
| 9 | HOOK2 | HOOK2 Entrez, Source | hook homolog 2 (Drosophila) | 1492 | 0.114 | 0.0070 | No |
| 10 | WDR23 | WDR23 Entrez, Source | WD repeat domain 23 | 1544 | 0.111 | 0.0124 | No |
| 11 | NDUFS1 | NDUFS1 Entrez, Source | NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) | 1817 | 0.101 | 0.0003 | No |
| 12 | SNIP | SNIP Entrez, Source | - | 2122 | 0.090 | -0.0151 | No |
| 13 | STC2 | STC2 Entrez, Source | stanniocalcin 2 | 2436 | 0.082 | -0.0320 | No |
| 14 | FGF5 | FGF5 Entrez, Source | fibroblast growth factor 5 | 2656 | 0.075 | -0.0423 | No |
| 15 | GNAS | GNAS Entrez, Source | GNAS complex locus | 2815 | 0.071 | -0.0483 | No |
| 16 | RAPGEF3 | RAPGEF3 Entrez, Source | Rap guanine nucleotide exchange factor (GEF) 3 | 2860 | 0.070 | -0.0458 | No |
| 17 | HMGN2 | HMGN2 Entrez, Source | high-mobility group nucleosomal binding domain 2 | 2936 | 0.068 | -0.0458 | No |
| 18 | WDR20 | WDR20 Entrez, Source | WD repeat domain 20 | 3021 | 0.066 | -0.0466 | No |
| 19 | WBP2 | WBP2 Entrez, Source | WW domain binding protein 2 | 3407 | 0.058 | -0.0708 | No |
| 20 | NFX1 | NFX1 Entrez, Source | nuclear transcription factor, X-box binding 1 | 3696 | 0.052 | -0.0883 | No |
| 21 | SEPT3 | SEPT3 Entrez, Source | septin 3 | 4226 | 0.042 | -0.1247 | No |
| 22 | ATP7B | ATP7B Entrez, Source | ATPase, Cu++ transporting, beta polypeptide | 4291 | 0.041 | -0.1260 | No |
| 23 | ELK1 | ELK1 Entrez, Source | ELK1, member of ETS oncogene family | 4490 | 0.038 | -0.1378 | No |
| 24 | TAC1 | TAC1 Entrez, Source | tachykinin, precursor 1 (substance K, substance P, neurokinin 1, neurokinin 2, neuromedin L, neurokinin alpha, neuropeptide K, neuropeptide gamma) | 4517 | 0.038 | -0.1367 | No |
| 25 | HPGD | HPGD Entrez, Source | hydroxyprostaglandin dehydrogenase 15-(NAD) | 4803 | 0.032 | -0.1555 | No |
| 26 | HNRPDL | HNRPDL Entrez, Source | heterogeneous nuclear ribonucleoprotein D-like | 4822 | 0.032 | -0.1542 | No |
| 27 | VPS16 | VPS16 Entrez, Source | vacuolar protein sorting 16 (yeast) | 5010 | 0.029 | -0.1659 | No |
| 28 | AATF | AATF Entrez, Source | apoptosis antagonizing transcription factor | 5022 | 0.029 | -0.1643 | No |
| 29 | RNF12 | RNF12 Entrez, Source | ring finger protein 12 | 5044 | 0.028 | -0.1635 | No |
| 30 | CLN3 | CLN3 Entrez, Source | ceroid-lipofuscinosis, neuronal 3, juvenile (Batten, Spielmeyer-Vogt disease) | 5208 | 0.026 | -0.1736 | No |
| 31 | EGLN2 | EGLN2 Entrez, Source | egl nine homolog 2 (C. elegans) | 5283 | 0.025 | -0.1772 | No |
| 32 | CDH1 | CDH1 Entrez, Source | cadherin 1, type 1, E-cadherin (epithelial) | 5315 | 0.024 | -0.1775 | No |
| 33 | AKAP12 | AKAP12 Entrez, Source | A kinase (PRKA) anchor protein (gravin) 12 | 5319 | 0.024 | -0.1757 | No |
| 34 | GTF2A1 | GTF2A1 Entrez, Source | general transcription factor IIA, 1, 19/37kDa | 5328 | 0.024 | -0.1742 | No |
| 35 | EIF3S9 | EIF3S9 Entrez, Source | eukaryotic translation initiation factor 3, subunit 9 eta, 116kDa | 5432 | 0.023 | -0.1801 | No |
| 36 | MYST3 | MYST3 Entrez, Source | MYST histone acetyltransferase (monocytic leukemia) 3 | 5502 | 0.022 | -0.1835 | No |
| 37 | ATP6V1H | ATP6V1H Entrez, Source | ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H | 5858 | 0.016 | -0.2090 | No |
| 38 | APEX1 | APEX1 Entrez, Source | APEX nuclease (multifunctional DNA repair enzyme) 1 | 6037 | 0.014 | -0.2213 | No |
| 39 | HOXA1 | HOXA1 Entrez, Source | homeobox A1 | 6083 | 0.013 | -0.2236 | No |
| 40 | RAB3IL1 | RAB3IL1 Entrez, Source | RAB3A interacting protein (rabin3)-like 1 | 6226 | 0.011 | -0.2334 | No |
| 41 | PPM1A | PPM1A Entrez, Source | protein phosphatase 1A (formerly 2C), magnesium-dependent, alpha isoform | 6343 | 0.009 | -0.2414 | No |
| 42 | PER1 | PER1 Entrez, Source | period homolog 1 (Drosophila) | 6654 | 0.005 | -0.2644 | No |
| 43 | APLN | APLN Entrez, Source | apelin, AGTRL1 ligand | 6690 | 0.004 | -0.2668 | No |
| 44 | EIF4E | EIF4E Entrez, Source | eukaryotic translation initiation factor 4E | 6703 | 0.004 | -0.2673 | No |
| 45 | SYT13 | SYT13 Entrez, Source | synaptotagmin XIII | 6837 | 0.002 | -0.2772 | No |
| 46 | NR3C2 | NR3C2 Entrez, Source | nuclear receptor subfamily 3, group C, member 2 | 6857 | 0.002 | -0.2785 | No |
| 47 | NPM1 | NPM1 Entrez, Source | nucleophosmin (nucleolar phosphoprotein B23, numatrin) | 7074 | -0.001 | -0.2947 | No |
| 48 | RPS2 | RPS2 Entrez, Source | ribosomal protein S2 | 7135 | -0.002 | -0.2990 | No |
| 49 | LTBR | LTBR Entrez, Source | lymphotoxin beta receptor (TNFR superfamily, member 3) | 7154 | -0.002 | -0.3002 | No |
| 50 | EIF4G1 | EIF4G1 Entrez, Source | eukaryotic translation initiation factor 4 gamma, 1 | 7196 | -0.003 | -0.3030 | No |
| 51 | SYT3 | SYT3 Entrez, Source | synaptotagmin III | 7475 | -0.007 | -0.3234 | No |
| 52 | LZTS2 | LZTS2 Entrez, Source | leucine zipper, putative tumor suppressor 2 | 7831 | -0.013 | -0.3492 | No |
| 53 | RPL19 | RPL19 Entrez, Source | ribosomal protein L19 | 7963 | -0.014 | -0.3579 | No |
| 54 | UBE2B | UBE2B Entrez, Source | ubiquitin-conjugating enzyme E2B (RAD6 homolog) | 8205 | -0.018 | -0.3746 | No |
| 55 | EIF5A | EIF5A Entrez, Source | eukaryotic translation initiation factor 5A | 8430 | -0.022 | -0.3897 | No |
| 56 | CHD4 | CHD4 Entrez, Source | chromodomain helicase DNA binding protein 4 | 8491 | -0.023 | -0.3923 | No |
| 57 | EWSR1 | EWSR1 Entrez, Source | Ewing sarcoma breakpoint region 1 | 8920 | -0.031 | -0.4221 | No |
| 58 | VGF | VGF Entrez, Source | VGF nerve growth factor inducible | 8973 | -0.032 | -0.4233 | No |
| 59 | DNMT3A | DNMT3A Entrez, Source | DNA (cytosine-5-)-methyltransferase 3 alpha | 9156 | -0.035 | -0.4342 | No |
| 60 | HRH3 | HRH3 Entrez, Source | histamine receptor H3 | 9350 | -0.039 | -0.4455 | No |
| 61 | BAX | BAX Entrez, Source | BCL2-associated X protein | 9773 | -0.047 | -0.4735 | No |
| 62 | KLF4 | KLF4 Entrez, Source | Kruppel-like factor 4 (gut) | 9940 | -0.051 | -0.4817 | No |
| 63 | COMMD3 | COMMD3 Entrez, Source | COMM domain containing 3 | 10033 | -0.053 | -0.4843 | No |
| 64 | NEUROD2 | NEUROD2 Entrez, Source | neurogenic differentiation 2 | 10039 | -0.053 | -0.4803 | No |
| 65 | LAMP1 | LAMP1 Entrez, Source | lysosomal-associated membrane protein 1 | 10118 | -0.055 | -0.4816 | No |
| 66 | CNOT4 | CNOT4 Entrez, Source | CCR4-NOT transcription complex, subunit 4 | 10146 | -0.055 | -0.4790 | No |
| 67 | ILF3 | ILF3 Entrez, Source | interleukin enhancer binding factor 3, 90kDa | 10199 | -0.057 | -0.4782 | No |
| 68 | RNF44 | RNF44 Entrez, Source | ring finger protein 44 | 10288 | -0.059 | -0.4799 | No |
| 69 | DCTN4 | DCTN4 Entrez, Source | dynactin 4 (p62) | 10318 | -0.060 | -0.4771 | No |
| 70 | TRIB1 | TRIB1 Entrez, Source | tribbles homolog 1 (Drosophila) | 10667 | -0.069 | -0.4977 | No |
| 71 | FGF14 | FGF14 Entrez, Source | fibroblast growth factor 14 | 11120 | -0.083 | -0.5249 | No |
| 72 | NEUROD1 | NEUROD1 Entrez, Source | neurogenic differentiation 1 | 11288 | -0.090 | -0.5300 | Yes |
| 73 | ARL3 | ARL3 Entrez, Source | ADP-ribosylation factor-like 3 | 11355 | -0.093 | -0.5273 | Yes |
| 74 | SLC26A2 | SLC26A2 Entrez, Source | solute carrier family 26 (sulfate transporter), member 2 | 11449 | -0.097 | -0.5261 | Yes |
| 75 | GNB2 | GNB2 Entrez, Source | guanine nucleotide binding protein (G protein), beta polypeptide 2 | 11499 | -0.099 | -0.5216 | Yes |
| 76 | DUSP7 | DUSP7 Entrez, Source | dual specificity phosphatase 7 | 11512 | -0.100 | -0.5141 | Yes |
| 77 | IGF2R | IGF2R Entrez, Source | insulin-like growth factor 2 receptor | 11527 | -0.101 | -0.5068 | Yes |
| 78 | RGL2 | RGL2 Entrez, Source | ral guanine nucleotide dissociation stimulator-like 2 | 11681 | -0.108 | -0.5093 | Yes |
| 79 | MAX | MAX Entrez, Source | MYC associated factor X | 11963 | -0.122 | -0.5203 | Yes |
| 80 | JUP | JUP Entrez, Source | junction plakoglobin | 12003 | -0.125 | -0.5128 | Yes |
| 81 | ADAM10 | ADAM10 Entrez, Source | ADAM metallopeptidase domain 10 | 12061 | -0.129 | -0.5064 | Yes |
| 82 | GIT1 | GIT1 Entrez, Source | G protein-coupled receptor kinase interactor 1 | 12064 | -0.129 | -0.4958 | Yes |
| 83 | NRAS | NRAS Entrez, Source | neuroblastoma RAS viral (v-ras) oncogene homolog | 12113 | -0.132 | -0.4884 | Yes |
| 84 | RAMP2 | RAMP2 Entrez, Source | receptor (calcitonin) activity modifying protein 2 | 12236 | -0.143 | -0.4857 | Yes |
| 85 | HEXA | HEXA Entrez, Source | hexosaminidase A (alpha polypeptide) | 12459 | -0.165 | -0.4887 | Yes |
| 86 | VLDLR | VLDLR Entrez, Source | very low density lipoprotein receptor | 12472 | -0.167 | -0.4757 | Yes |
| 87 | HIF1A | HIF1A Entrez, Source | hypoxia-inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) | 12528 | -0.175 | -0.4653 | Yes |
| 88 | PICALM | PICALM Entrez, Source | phosphatidylinositol binding clathrin assembly protein | 12640 | -0.193 | -0.4576 | Yes |
| 89 | SGK | SGK Entrez, Source | serum/glucocorticoid regulated kinase | 12735 | -0.210 | -0.4471 | Yes |
| 90 | SNN | SNN Entrez, Source | stannin | 12747 | -0.213 | -0.4302 | Yes |
| 91 | STX6 | STX6 Entrez, Source | syntaxin 6 | 12878 | -0.244 | -0.4196 | Yes |
| 92 | WEE1 | WEE1 Entrez, Source | WEE1 homolog (S. pombe) | 12929 | -0.262 | -0.4015 | Yes |
| 93 | PTMA | PTMA Entrez, Source | prothymosin, alpha (gene sequence 28) | 12957 | -0.275 | -0.3806 | Yes |
| 94 | WDR1 | WDR1 Entrez, Source | WD repeat domain 1 | 12973 | -0.280 | -0.3584 | Yes |
| 95 | HNRPD | HNRPD Entrez, Source | heterogeneous nuclear ribonucleoprotein D (AU-rich element RNA binding protein 1, 37kDa) | 13048 | -0.319 | -0.3374 | Yes |
| 96 | RHOQ | RHOQ Entrez, Source | ras homolog gene family, member Q | 13120 | -0.369 | -0.3120 | Yes |
| 97 | SCHIP1 | SCHIP1 Entrez, Source | schwannomin interacting protein 1 | 13180 | -0.438 | -0.2799 | Yes |
| 98 | RAB31 | RAB31 Entrez, Source | RAB31, member RAS oncogene family | 13187 | -0.451 | -0.2427 | Yes |
| 99 | SLC31A2 | SLC31A2 Entrez, Source | solute carrier family 31 (copper transporters), member 2 | 13228 | -0.508 | -0.2034 | Yes |
| 100 | BDNF | BDNF Entrez, Source | brain-derived neurotrophic factor | 13242 | -0.532 | -0.1600 | Yes |
| 101 | STMN1 | STMN1 Entrez, Source | stathmin 1/oncoprotein 18 | 13302 | -0.755 | -0.1015 | Yes |
| 102 | BHLHB3 | BHLHB3 Entrez, Source | basic helix-loop-helix domain containing, class B, 3 | 13335 | -1.251 | 0.0005 | Yes |