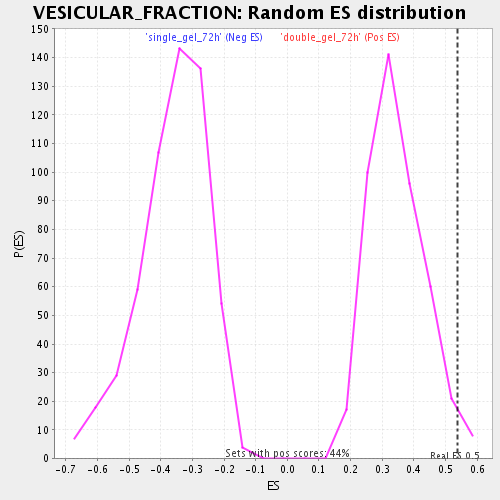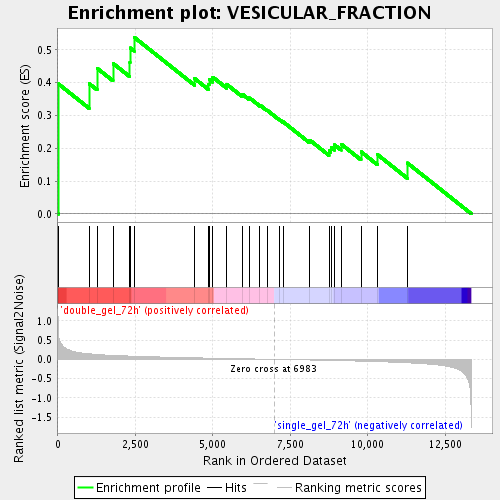
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_72h_versus_single_gel_72h.class.cls #double_gel_72h_versus_single_gel_72h_repos |
| Phenotype | class.cls#double_gel_72h_versus_single_gel_72h_repos |
| Upregulated in class | double_gel_72h |
| GeneSet | VESICULAR_FRACTION |
| Enrichment Score (ES) | 0.5381966 |
| Normalized Enrichment Score (NES) | 1.5559884 |
| Nominal p-value | 0.024830699 |
| FDR q-value | 0.0865733 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | AADAC | AADAC Entrez, Source | arylacetamide deacetylase (esterase) | 11 | 0.758 | 0.3963 | Yes |
| 2 | APOB | APOB Entrez, Source | apolipoprotein B (including Ag(x) antigen) | 1009 | 0.144 | 0.3968 | Yes |
| 3 | COMT | COMT Entrez, Source | catechol-O-methyltransferase | 1267 | 0.126 | 0.4435 | Yes |
| 4 | SLC35B1 | SLC35B1 Entrez, Source | solute carrier family 35, member B1 | 1780 | 0.102 | 0.4584 | Yes |
| 5 | FMO3 | FMO3 Entrez, Source | flavin containing monooxygenase 3 | 2322 | 0.084 | 0.4620 | Yes |
| 6 | IRS1 | IRS1 Entrez, Source | insulin receptor substrate 1 | 2329 | 0.084 | 0.5057 | Yes |
| 7 | SSR3 | SSR3 Entrez, Source | signal sequence receptor, gamma (translocon-associated protein gamma) | 2461 | 0.081 | 0.5382 | Yes |
| 8 | STCH | STCH Entrez, Source | stress 70 protein chaperone, microsome-associated, 60kDa | 4413 | 0.039 | 0.4123 | No |
| 9 | DHRS9 | DHRS9 Entrez, Source | dehydrogenase/reductase (SDR family) member 9 | 4865 | 0.031 | 0.3949 | No |
| 10 | PTGS1 | PTGS1 Entrez, Source | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | 4887 | 0.031 | 0.4096 | No |
| 11 | HSD3B1 | HSD3B1 Entrez, Source | hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 1 | 4997 | 0.029 | 0.4167 | No |
| 12 | COPA | COPA Entrez, Source | coatomer protein complex, subunit alpha | 5449 | 0.022 | 0.3946 | No |
| 13 | ALS2 | ALS2 Entrez, Source | amyotrophic lateral sclerosis 2 (juvenile) | 5959 | 0.015 | 0.3641 | No |
| 14 | TMED10 | TMED10 Entrez, Source | transmembrane emp24-like trafficking protein 10 (yeast) | 6185 | 0.012 | 0.3533 | No |
| 15 | STX2 | STX2 Entrez, Source | syntaxin 2 | 6520 | 0.006 | 0.3314 | No |
| 16 | CYP2C9 | CYP2C9 Entrez, Source | cytochrome P450, family 2, subfamily C, polypeptide 9 | 6753 | 0.003 | 0.3156 | No |
| 17 | TMED2 | TMED2 Entrez, Source | transmembrane emp24 domain trafficking protein 2 | 7149 | -0.002 | 0.2872 | No |
| 18 | GBA2 | GBA2 Entrez, Source | glucosidase, beta (bile acid) 2 | 7269 | -0.004 | 0.2804 | No |
| 19 | LTC4S | LTC4S Entrez, Source | leukotriene C4 synthase | 8127 | -0.017 | 0.2248 | No |
| 20 | HSD17B3 | HSD17B3 Entrez, Source | hydroxysteroid (17-beta) dehydrogenase 3 | 8751 | -0.027 | 0.1923 | No |
| 21 | POMGNT1 | POMGNT1 Entrez, Source | protein O-linked mannose beta1,2-N-acetylglucosaminyltransferase | 8813 | -0.028 | 0.2026 | No |
| 22 | ERO1L | ERO1L Entrez, Source | ERO1-like (S. cerevisiae) | 8926 | -0.031 | 0.2103 | No |
| 23 | VCP | VCP Entrez, Source | valosin-containing protein | 9139 | -0.035 | 0.2125 | No |
| 24 | ATP2A2 | ATP2A2 Entrez, Source | ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 | 9784 | -0.048 | 0.1891 | No |
| 25 | CYP3A4 | CYP3A4 Entrez, Source | cytochrome P450, family 3, subfamily A, polypeptide 4 | 10311 | -0.059 | 0.1807 | No |
| 26 | STS | STS Entrez, Source | steroid sulfatase (microsomal), arylsulfatase C, isozyme S | 11277 | -0.089 | 0.1551 | No |