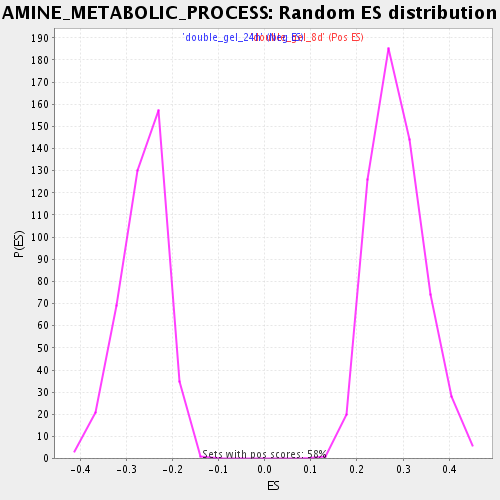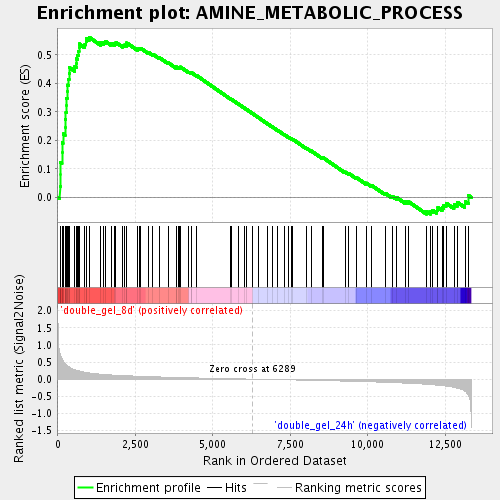
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_double_gel_24h.class.cls #double_gel_8d_versus_double_gel_24h_repos |
| Phenotype | class.cls#double_gel_8d_versus_double_gel_24h_repos |
| Upregulated in class | double_gel_8d |
| GeneSet | AMINE_METABOLIC_PROCESS |
| Enrichment Score (ES) | 0.5614974 |
| Normalized Enrichment Score (NES) | 1.9596951 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0074743084 |
| FWER p-Value | 0.269 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | ST3GAL2 | ST3GAL2 Entrez, Source | ST3 beta-galactoside alpha-2,3-sialyltransferase 2 | 67 | 0.744 | 0.0402 | Yes |
| 2 | GOT1 | GOT1 Entrez, Source | glutamic-oxaloacetic transaminase 1, soluble (aspartate aminotransferase 1) | 76 | 0.697 | 0.0820 | Yes |
| 3 | BBOX1 | BBOX1 Entrez, Source | butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 | 78 | 0.695 | 0.1242 | Yes |
| 4 | PAH | PAH Entrez, Source | phenylalanine hydroxylase | 133 | 0.597 | 0.1565 | Yes |
| 5 | HGD | HGD Entrez, Source | homogentisate 1,2-dioxygenase (homogentisate oxidase) | 138 | 0.591 | 0.1922 | Yes |
| 6 | SULT1A1 | SULT1A1 Entrez, Source | sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 | 166 | 0.540 | 0.2230 | Yes |
| 7 | FAH | FAH Entrez, Source | fumarylacetoacetate hydrolase (fumarylacetoacetase) | 232 | 0.457 | 0.2459 | Yes |
| 8 | SULT1B1 | SULT1B1 Entrez, Source | sulfotransferase family, cytosolic, 1B, member 1 | 238 | 0.454 | 0.2732 | Yes |
| 9 | BCKDHA | BCKDHA Entrez, Source | branched chain keto acid dehydrogenase E1, alpha polypeptide | 258 | 0.431 | 0.2980 | Yes |
| 10 | GGT1 | GGT1 Entrez, Source | gamma-glutamyltransferase 1 | 271 | 0.423 | 0.3228 | Yes |
| 11 | DDAH1 | DDAH1 Entrez, Source | dimethylarginine dimethylaminohydrolase 1 | 276 | 0.416 | 0.3478 | Yes |
| 12 | ALDH6A1 | ALDH6A1 Entrez, Source | aldehyde dehydrogenase 6 family, member A1 | 300 | 0.402 | 0.3705 | Yes |
| 13 | ARG1 | ARG1 Entrez, Source | arginase, liver | 307 | 0.397 | 0.3943 | Yes |
| 14 | SLC25A15 | SLC25A15 Entrez, Source | solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 15 | 338 | 0.371 | 0.4146 | Yes |
| 15 | MAT1A | MAT1A Entrez, Source | methionine adenosyltransferase I, alpha | 362 | 0.357 | 0.4346 | Yes |
| 16 | SLC3A1 | SLC3A1 Entrez, Source | 1 | 373 | 0.349 | 0.4551 | Yes |
| 17 | GLS2 | GLS2 Entrez, Source | glutaminase 2 (liver, mitochondrial) | 543 | 0.271 | 0.4588 | Yes |
| 18 | ALDH5A1 | ALDH5A1 Entrez, Source | aldehyde dehydrogenase 5 family, member A1 (succinate-semialdehyde dehydrogenase) | 595 | 0.258 | 0.4707 | Yes |
| 19 | BAAT | BAAT Entrez, Source | bile acid Coenzyme A: amino acid N-acyltransferase (glycine N-choloyltransferase) | 602 | 0.257 | 0.4858 | Yes |
| 20 | GUSB | GUSB Entrez, Source | glucuronidase, beta | 637 | 0.247 | 0.4983 | Yes |
| 21 | MCCC2 | MCCC2 Entrez, Source | methylcrotonoyl-Coenzyme A carboxylase 2 (beta) | 663 | 0.243 | 0.5112 | Yes |
| 22 | ADI1 | ADI1 Entrez, Source | acireductone dioxygenase 1 | 682 | 0.239 | 0.5244 | Yes |
| 23 | CDO1 | CDO1 Entrez, Source | cysteine dioxygenase, type I | 689 | 0.237 | 0.5383 | Yes |
| 24 | BCKDK | BCKDK Entrez, Source | branched chain ketoacid dehydrogenase kinase | 869 | 0.200 | 0.5370 | Yes |
| 25 | BCKDHB | BCKDHB Entrez, Source | branched chain keto acid dehydrogenase E1, beta polypeptide (maple syrup urine disease) | 906 | 0.195 | 0.5461 | Yes |
| 26 | TGFB2 | TGFB2 Entrez, Source | transforming growth factor, beta 2 | 916 | 0.193 | 0.5572 | Yes |
| 27 | SDS | SDS Entrez, Source | serine dehydratase | 1007 | 0.182 | 0.5615 | Yes |
| 28 | MSRA | MSRA Entrez, Source | methionine sulfoxide reductase A | 1379 | 0.144 | 0.5423 | No |
| 29 | PTS | PTS Entrez, Source | 6-pyruvoyltetrahydropterin synthase | 1478 | 0.137 | 0.5432 | No |
| 30 | GAMT | GAMT Entrez, Source | guanidinoacetate N-methyltransferase | 1535 | 0.132 | 0.5470 | No |
| 31 | SLC7A2 | SLC7A2 Entrez, Source | solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 | 1713 | 0.122 | 0.5411 | No |
| 32 | DDAH2 | DDAH2 Entrez, Source | dimethylarginine dimethylaminohydrolase 2 | 1832 | 0.115 | 0.5392 | No |
| 33 | SLC5A7 | SLC5A7 Entrez, Source | solute carrier family 5 (choline transporter), member 7 | 1871 | 0.113 | 0.5432 | No |
| 34 | ASPA | ASPA Entrez, Source | aspartoacylase (Canavan disease) | 2090 | 0.104 | 0.5331 | No |
| 35 | HDC | HDC Entrez, Source | histidine decarboxylase | 2155 | 0.101 | 0.5344 | No |
| 36 | CHST3 | CHST3 Entrez, Source | carbohydrate (chondroitin 6) sulfotransferase 3 | 2214 | 0.099 | 0.5361 | No |
| 37 | AMT | AMT Entrez, Source | aminomethyltransferase | 2221 | 0.098 | 0.5416 | No |
| 38 | NFS1 | NFS1 Entrez, Source | NFS1 nitrogen fixation 1 (S. cerevisiae) | 2570 | 0.085 | 0.5205 | No |
| 39 | GNS | GNS Entrez, Source | glucosamine (N-acetyl)-6-sulfatase (Sanfilippo disease IIID) | 2618 | 0.083 | 0.5220 | No |
| 40 | COLQ | COLQ Entrez, Source | collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase | 2670 | 0.081 | 0.5231 | No |
| 41 | B4GALT7 | B4GALT7 Entrez, Source | xylosylprotein beta 1,4-galactosyltransferase, polypeptide 7 (galactosyltransferase I) | 2931 | 0.073 | 0.5079 | No |
| 42 | GNE | GNE Entrez, Source | glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase | 3063 | 0.069 | 0.5023 | No |
| 43 | CHIA | CHIA Entrez, Source | chitinase, acidic | 3265 | 0.064 | 0.4910 | No |
| 44 | SLC7A9 | SLC7A9 Entrez, Source | solute carrier family 7 (cationic amino acid transporter, y+ system), member 9 | 3559 | 0.056 | 0.4723 | No |
| 45 | FARS2 | FARS2 Entrez, Source | phenylalanine-tRNA synthetase 2 (mitochondrial) | 3826 | 0.050 | 0.4552 | No |
| 46 | SCLY | SCLY Entrez, Source | selenocysteine lyase | 3833 | 0.049 | 0.4577 | No |
| 47 | GCSH | GCSH Entrez, Source | glycine cleavage system protein H (aminomethyl carrier) | 3887 | 0.048 | 0.4566 | No |
| 48 | BCAT1 | BCAT1 Entrez, Source | branched chain aminotransferase 1, cytosolic | 3938 | 0.046 | 0.4557 | No |
| 49 | CHST12 | CHST12 Entrez, Source | carbohydrate (chondroitin 4) sulfotransferase 12 | 3952 | 0.046 | 0.4575 | No |
| 50 | CHST1 | CHST1 Entrez, Source | carbohydrate (keratan sulfate Gal-6) sulfotransferase 1 | 4227 | 0.039 | 0.4392 | No |
| 51 | GLUD1 | GLUD1 Entrez, Source | glutamate dehydrogenase 1 | 4297 | 0.038 | 0.4362 | No |
| 52 | XYLT1 | XYLT1 Entrez, Source | xylosyltransferase I | 4309 | 0.037 | 0.4377 | No |
| 53 | EXTL2 | EXTL2 Entrez, Source | exostoses (multiple)-like 2 | 4481 | 0.034 | 0.4269 | No |
| 54 | GGTLA1 | GGTLA1 Entrez, Source | gamma-glutamyltransferase-like activity 1 | 5579 | 0.013 | 0.3449 | No |
| 55 | AOC3 | AOC3 Entrez, Source | amine oxidase, copper containing 3 (vascular adhesion protein 1) | 5594 | 0.013 | 0.3446 | No |
| 56 | SLC7A8 | SLC7A8 Entrez, Source | solute carrier family 7 (cationic amino acid transporter, y+ system), member 8 | 5837 | 0.008 | 0.3268 | No |
| 57 | GALNT5 | GALNT5 Entrez, Source | UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5) | 6021 | 0.005 | 0.3133 | No |
| 58 | ASRGL1 | ASRGL1 Entrez, Source | asparaginase like 1 | 6099 | 0.003 | 0.3077 | No |
| 59 | ATF4 | ATF4 Entrez, Source | activating transcription factor 4 (tax-responsive enhancer element B67) | 6295 | -0.000 | 0.2930 | No |
| 60 | HPD | HPD Entrez, Source | 4-hydroxyphenylpyruvate dioxygenase | 6481 | -0.003 | 0.2792 | No |
| 61 | ASL | ASL Entrez, Source | argininosuccinate lyase | 6773 | -0.008 | 0.2578 | No |
| 62 | SLC6A6 | SLC6A6 Entrez, Source | solute carrier family 6 (neurotransmitter transporter, taurine), member 6 | 6923 | -0.011 | 0.2472 | No |
| 63 | SPHK1 | SPHK1 Entrez, Source | sphingosine kinase 1 | 7090 | -0.014 | 0.2355 | No |
| 64 | DHPS | DHPS Entrez, Source | deoxyhypusine synthase | 7301 | -0.018 | 0.2208 | No |
| 65 | QDPR | QDPR Entrez, Source | quinoid dihydropteridine reductase | 7441 | -0.021 | 0.2115 | No |
| 66 | GSS | GSS Entrez, Source | glutathione synthetase | 7538 | -0.022 | 0.2056 | No |
| 67 | NANP | NANP Entrez, Source | N-acetylneuraminic acid phosphatase | 7570 | -0.023 | 0.2047 | No |
| 68 | OAZ1 | OAZ1 Entrez, Source | ornithine decarboxylase antizyme 1 | 8028 | -0.032 | 0.1721 | No |
| 69 | NAGK | NAGK Entrez, Source | N-acetylglucosamine kinase | 8172 | -0.035 | 0.1634 | No |
| 70 | DIO2 | DIO2 Entrez, Source | deiodinase, iodothyronine, type II | 8533 | -0.042 | 0.1388 | No |
| 71 | ST3GAL6 | ST3GAL6 Entrez, Source | ST3 beta-galactoside alpha-2,3-sialyltransferase 6 | 8557 | -0.043 | 0.1397 | No |
| 72 | GAD1 | GAD1 Entrez, Source | glutamate decarboxylase 1 (brain, 67kDa) | 9270 | -0.057 | 0.0895 | No |
| 73 | HPRT1 | HPRT1 Entrez, Source | hypoxanthine phosphoribosyltransferase 1 (Lesch-Nyhan syndrome) | 9384 | -0.060 | 0.0846 | No |
| 74 | GOT2 | GOT2 Entrez, Source | glutamic-oxaloacetic transaminase 2, mitochondrial (aspartate aminotransferase 2) | 9641 | -0.066 | 0.0693 | No |
| 75 | YARS | YARS Entrez, Source | tyrosyl-tRNA synthetase | 9947 | -0.075 | 0.0508 | No |
| 76 | P4HB | P4HB Entrez, Source | procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), beta polypeptide | 10130 | -0.080 | 0.0420 | No |
| 77 | INDO | INDO Entrez, Source | indoleamine-pyrrole 2,3 dioxygenase | 10558 | -0.092 | 0.0153 | No |
| 78 | GATM | GATM Entrez, Source | glycine amidinotransferase (L-arginine:glycine amidinotransferase) | 10782 | -0.100 | 0.0046 | No |
| 79 | KARS | KARS Entrez, Source | lysyl-tRNA synthetase | 10927 | -0.106 | 0.0002 | No |
| 80 | GAD2 | GAD2 Entrez, Source | glutamate decarboxylase 2 (pancreatic islets and brain, 65kDa) | 11204 | -0.116 | -0.0136 | No |
| 81 | PLOD1 | PLOD1 Entrez, Source | procollagen-lysine 1, 2-oxoglutarate 5-dioxygenase 1 | 11305 | -0.121 | -0.0138 | No |
| 82 | SLC7A7 | SLC7A7 Entrez, Source | solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 | 11894 | -0.150 | -0.0490 | No |
| 83 | DARS | DARS Entrez, Source | aspartyl-tRNA synthetase | 12016 | -0.158 | -0.0485 | No |
| 84 | WARS | WARS Entrez, Source | tryptophanyl-tRNA synthetase | 12088 | -0.163 | -0.0439 | No |
| 85 | AARS | AARS Entrez, Source | alanyl-tRNA synthetase | 12234 | -0.174 | -0.0443 | No |
| 86 | SLC35A3 | SLC35A3 Entrez, Source | solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3 | 12262 | -0.176 | -0.0356 | No |
| 87 | SCYE1 | SCYE1 Entrez, Source | small inducible cytokine subfamily E, member 1 (endothelial monocyte-activating) | 12401 | -0.188 | -0.0345 | No |
| 88 | SULT1C2 | SULT1C2 Entrez, Source | sulfotransferase family, cytosolic, 1C, member 2 | 12449 | -0.193 | -0.0263 | No |
| 89 | SMS | SMS Entrez, Source | spermine synthase | 12527 | -0.202 | -0.0198 | No |
| 90 | GCLM | GCLM Entrez, Source | glutamate-cysteine ligase, modifier subunit | 12784 | -0.238 | -0.0247 | No |
| 91 | SLC7A5 | SLC7A5 Entrez, Source | solute carrier family 7 (cationic amino acid transporter, y+ system), member 5 | 12884 | -0.262 | -0.0162 | No |
| 92 | UGDH | UGDH Entrez, Source | UDP-glucose dehydrogenase | 13138 | -0.352 | -0.0139 | No |
| 93 | GCLC | GCLC Entrez, Source | glutamate-cysteine ligase, catalytic subunit | 13249 | -0.480 | 0.0070 | No |