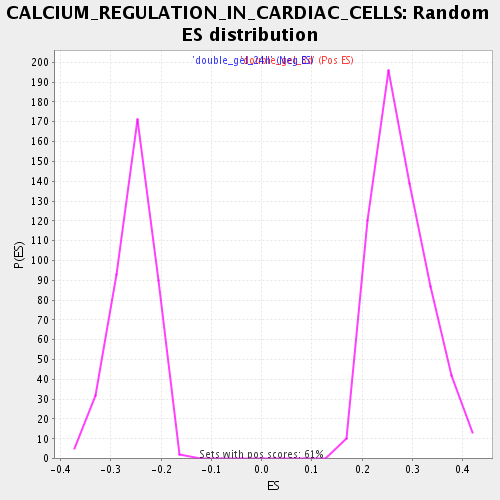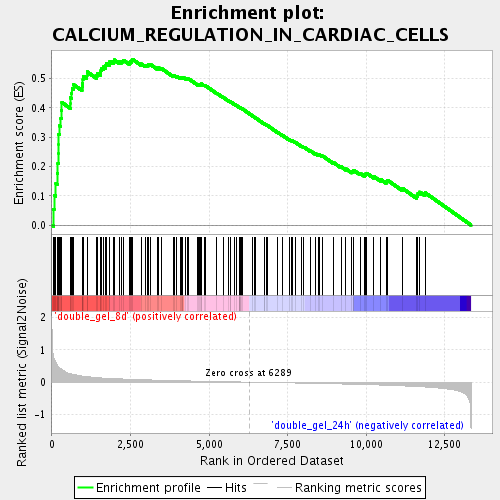
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_double_gel_24h.class.cls #double_gel_8d_versus_double_gel_24h_repos |
| Phenotype | class.cls#double_gel_8d_versus_double_gel_24h_repos |
| Upregulated in class | double_gel_8d |
| GeneSet | CALCIUM_REGULATION_IN_CARDIAC_CELLS |
| Enrichment Score (ES) | 0.5662463 |
| Normalized Enrichment Score (NES) | 2.0508406 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0028363036 |
| FWER p-Value | 0.058 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | RGS2 | RGS2 Entrez, Source | regulator of G-protein signalling 2, 24kDa | 50 | 0.810 | 0.0534 | Yes |
| 2 | RGS1 | RGS1 Entrez, Source | regulator of G-protein signalling 1 | 73 | 0.715 | 0.1022 | Yes |
| 3 | RGS5 | RGS5 Entrez, Source | regulator of G-protein signalling 5 | 125 | 0.616 | 0.1419 | Yes |
| 4 | GJB5 | GJB5 Entrez, Source | gap junction protein, beta 5 (connexin 31.1) | 171 | 0.530 | 0.1759 | Yes |
| 5 | GRK5 | GRK5 Entrez, Source | G protein-coupled receptor kinase 5 | 188 | 0.507 | 0.2105 | Yes |
| 6 | RGS16 | RGS16 Entrez, Source | regulator of G-protein signalling 16 | 205 | 0.482 | 0.2434 | Yes |
| 7 | GJB3 | GJB3 Entrez, Source | gap junction protein, beta 3, 31kDa (connexin 31) | 210 | 0.476 | 0.2767 | Yes |
| 8 | PRKAR2B | PRKAR2B Entrez, Source | protein kinase, cAMP-dependent, regulatory, type II, beta | 214 | 0.474 | 0.3100 | Yes |
| 9 | RGS10 | RGS10 Entrez, Source | regulator of G-protein signalling 10 | 247 | 0.443 | 0.3388 | Yes |
| 10 | ITPR1 | ITPR1 Entrez, Source | inositol 1,4,5-triphosphate receptor, type 1 | 282 | 0.413 | 0.3654 | Yes |
| 11 | RGS3 | RGS3 Entrez, Source | regulator of G-protein signalling 3 | 301 | 0.402 | 0.3925 | Yes |
| 12 | GJA1 | GJA1 Entrez, Source | gap junction protein, alpha 1, 43kDa (connexin 43) | 316 | 0.387 | 0.4187 | Yes |
| 13 | PKIA | PKIA Entrez, Source | protein kinase (cAMP-dependent, catalytic) inhibitor alpha | 588 | 0.260 | 0.4166 | Yes |
| 14 | PKIB | PKIB Entrez, Source | protein kinase (cAMP-dependent, catalytic) inhibitor beta | 592 | 0.258 | 0.4346 | Yes |
| 15 | GJB2 | GJB2 Entrez, Source | gap junction protein, beta 2, 26kDa (connexin 26) | 628 | 0.250 | 0.4496 | Yes |
| 16 | RGS4 | RGS4 Entrez, Source | regulator of G-protein signalling 4 | 639 | 0.247 | 0.4662 | Yes |
| 17 | GJB1 | GJB1 Entrez, Source | gap junction protein, beta 1, 32kDa (connexin 32, Charcot-Marie-Tooth neuropathy, X-linked) | 690 | 0.237 | 0.4792 | Yes |
| 18 | FXYD2 | FXYD2 Entrez, Source | FXYD domain containing ion transport regulator 2 | 960 | 0.187 | 0.4721 | Yes |
| 19 | ATP1B1 | ATP1B1 Entrez, Source | ATPase, Na+/K+ transporting, beta 1 polypeptide | 973 | 0.186 | 0.4843 | Yes |
| 20 | ADRB2 | ADRB2 Entrez, Source | adrenergic, beta-2-, receptor, surface | 987 | 0.184 | 0.4963 | Yes |
| 21 | PLN | PLN Entrez, Source | phospholamban | 994 | 0.183 | 0.5088 | Yes |
| 22 | PRKAR1B | PRKAR1B Entrez, Source | protein kinase, cAMP-dependent, regulatory, type I, beta | 1117 | 0.169 | 0.5115 | Yes |
| 23 | PKIG | PKIG Entrez, Source | protein kinase (cAMP-dependent, catalytic) inhibitor gamma | 1121 | 0.169 | 0.5232 | Yes |
| 24 | PRKCA | PRKCA Entrez, Source | protein kinase C, alpha | 1423 | 0.141 | 0.5104 | Yes |
| 25 | YWHAQ | YWHAQ Entrez, Source | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide | 1447 | 0.139 | 0.5184 | Yes |
| 26 | ADCY5 | ADCY5 Entrez, Source | adenylate cyclase 5 | 1541 | 0.132 | 0.5207 | Yes |
| 27 | ADRA1D | ADRA1D Entrez, Source | adrenergic, alpha-1D-, receptor | 1559 | 0.131 | 0.5287 | Yes |
| 28 | GNAI2 | GNAI2 Entrez, Source | guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2 | 1585 | 0.129 | 0.5359 | Yes |
| 29 | ADCY6 | ADCY6 Entrez, Source | adenylate cyclase 6 | 1649 | 0.125 | 0.5400 | Yes |
| 30 | ANXA6 | ANXA6 Entrez, Source | annexin A6 | 1714 | 0.122 | 0.5437 | Yes |
| 31 | GNAQ | GNAQ Entrez, Source | guanine nucleotide binding protein (G protein), q polypeptide | 1722 | 0.122 | 0.5518 | Yes |
| 32 | ATP2A3 | ATP2A3 Entrez, Source | ATPase, Ca++ transporting, ubiquitous | 1837 | 0.115 | 0.5513 | Yes |
| 33 | CACNA1E | CACNA1E Entrez, Source | calcium channel, voltage-dependent, alpha 1E subunit | 1843 | 0.115 | 0.5590 | Yes |
| 34 | ATP1A4 | ATP1A4 Entrez, Source | ATPase, Na+/K+ transporting, alpha 4 polypeptide | 1959 | 0.109 | 0.5580 | Yes |
| 35 | CALM3 | CALM3 Entrez, Source | calmodulin 3 (phosphorylase kinase, delta) | 1983 | 0.109 | 0.5639 | Yes |
| 36 | ADRB3 | ADRB3 Entrez, Source | adrenergic, beta-3-, receptor | 2147 | 0.102 | 0.5588 | Yes |
| 37 | ARRB1 | ARRB1 Entrez, Source | arrestin, beta 1 | 2228 | 0.098 | 0.5597 | Yes |
| 38 | RGS7 | RGS7 Entrez, Source | regulator of G-protein signalling 7 | 2271 | 0.097 | 0.5633 | Yes |
| 39 | GNB5 | GNB5 Entrez, Source | guanine nucleotide binding protein (G protein), beta 5 | 2462 | 0.089 | 0.5552 | Yes |
| 40 | PEA15 | PEA15 Entrez, Source | phosphoprotein enriched in astrocytes 15 | 2496 | 0.088 | 0.5590 | Yes |
| 41 | PRKCH | PRKCH Entrez, Source | protein kinase C, eta | 2545 | 0.086 | 0.5614 | Yes |
| 42 | ITPR3 | ITPR3 Entrez, Source | inositol 1,4,5-triphosphate receptor, type 3 | 2561 | 0.085 | 0.5662 | Yes |
| 43 | GNB3 | GNB3 Entrez, Source | guanine nucleotide binding protein (G protein), beta polypeptide 3 | 2846 | 0.076 | 0.5501 | No |
| 44 | GNG3 | GNG3 Entrez, Source | guanine nucleotide binding protein (G protein), gamma 3 | 2978 | 0.072 | 0.5453 | No |
| 45 | GNB4 | GNB4 Entrez, Source | guanine nucleotide binding protein (G protein), beta polypeptide 4 | 3031 | 0.070 | 0.5463 | No |
| 46 | PRKAR2A | PRKAR2A Entrez, Source | protein kinase, cAMP-dependent, regulatory, type II, alpha | 3077 | 0.069 | 0.5478 | No |
| 47 | ATP2B3 | ATP2B3 Entrez, Source | ATPase, Ca++ transporting, plasma membrane 3 | 3129 | 0.068 | 0.5487 | No |
| 48 | RGS6 | RGS6 Entrez, Source | regulator of G-protein signalling 6 | 3368 | 0.061 | 0.5350 | No |
| 49 | ATP1B2 | ATP1B2 Entrez, Source | ATPase, Na+/K+ transporting, beta 2 polypeptide | 3398 | 0.060 | 0.5371 | No |
| 50 | SLC8A1 | SLC8A1 Entrez, Source | solute carrier family 8 (sodium/calcium exchanger), member 1 | 3484 | 0.058 | 0.5347 | No |
| 51 | PRKCZ | PRKCZ Entrez, Source | protein kinase C, zeta | 3870 | 0.048 | 0.5090 | No |
| 52 | CACNB3 | CACNB3 Entrez, Source | calcium channel, voltage-dependent, beta 3 subunit | 3904 | 0.047 | 0.5099 | No |
| 53 | ARRB2 | ARRB2 Entrez, Source | arrestin, beta 2 | 3976 | 0.045 | 0.5077 | No |
| 54 | CHRM3 | CHRM3 Entrez, Source | cholinergic receptor, muscarinic 3 | 4080 | 0.043 | 0.5029 | No |
| 55 | CHRM2 | CHRM2 Entrez, Source | cholinergic receptor, muscarinic 2 | 4127 | 0.042 | 0.5024 | No |
| 56 | ADCY2 | ADCY2 Entrez, Source | adenylate cyclase 2 (brain) | 4158 | 0.041 | 0.5030 | No |
| 57 | GNG7 | GNG7 Entrez, Source | guanine nucleotide binding protein (G protein), gamma 7 | 4166 | 0.041 | 0.5053 | No |
| 58 | CAMK1 | CAMK1 Entrez, Source | calcium/calmodulin-dependent protein kinase I | 4262 | 0.038 | 0.5008 | No |
| 59 | CAMK2A | CAMK2A Entrez, Source | calcium/calmodulin-dependent protein kinase (CaM kinase) II alpha | 4303 | 0.037 | 0.5005 | No |
| 60 | KCNB1 | KCNB1 Entrez, Source | potassium voltage-gated channel, Shab-related subfamily, member 1 | 4356 | 0.037 | 0.4991 | No |
| 61 | CHRM4 | CHRM4 Entrez, Source | cholinergic receptor, muscarinic 4 | 4648 | 0.031 | 0.4793 | No |
| 62 | CAMK4 | CAMK4 Entrez, Source | calcium/calmodulin-dependent protein kinase IV | 4665 | 0.031 | 0.4803 | No |
| 63 | RYR1 | RYR1 Entrez, Source | ryanodine receptor 1 (skeletal) | 4695 | 0.030 | 0.4802 | No |
| 64 | ITPR2 | ITPR2 Entrez, Source | inositol 1,4,5-triphosphate receptor, type 2 | 4733 | 0.030 | 0.4795 | No |
| 65 | CACNA1D | CACNA1D Entrez, Source | calcium channel, voltage-dependent, L type, alpha 1D subunit | 4746 | 0.029 | 0.4806 | No |
| 66 | PRKAR1A | PRKAR1A Entrez, Source | protein kinase, cAMP-dependent, regulatory, type I, alpha (tissue specific extinguisher 1) | 4750 | 0.029 | 0.4824 | No |
| 67 | GJB6 | GJB6 Entrez, Source | gap junction protein, beta 6 (connexin 30) | 4849 | 0.027 | 0.4769 | No |
| 68 | CHRM5 | CHRM5 Entrez, Source | cholinergic receptor, muscarinic 5 | 4885 | 0.026 | 0.4762 | No |
| 69 | RGS19 | RGS19 Entrez, Source | regulator of G-protein signalling 19 | 5241 | 0.020 | 0.4507 | No |
| 70 | CACNA1B | CACNA1B Entrez, Source | calcium channel, voltage-dependent, L type, alpha 1B subunit | 5446 | 0.016 | 0.4364 | No |
| 71 | GNAI3 | GNAI3 Entrez, Source | guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 3 | 5631 | 0.012 | 0.4233 | No |
| 72 | PLCB3 | PLCB3 Entrez, Source | phospholipase C, beta 3 (phosphatidylinositol-specific) | 5675 | 0.011 | 0.4209 | No |
| 73 | GJA4 | GJA4 Entrez, Source | gap junction protein, alpha 4, 37kDa (connexin 37) | 5689 | 0.011 | 0.4207 | No |
| 74 | GNB2 | GNB2 Entrez, Source | guanine nucleotide binding protein (G protein), beta polypeptide 2 | 5801 | 0.009 | 0.4130 | No |
| 75 | RGS9 | RGS9 Entrez, Source | regulator of G-protein signalling 9 | 5873 | 0.008 | 0.4081 | No |
| 76 | CACNB1 | CACNB1 Entrez, Source | calcium channel, voltage-dependent, beta 1 subunit | 5959 | 0.006 | 0.4021 | No |
| 77 | CAMK2D | CAMK2D Entrez, Source | calcium/calmodulin-dependent protein kinase (CaM kinase) II delta | 5975 | 0.006 | 0.4014 | No |
| 78 | GRK4 | GRK4 Entrez, Source | G protein-coupled receptor kinase 4 | 5984 | 0.005 | 0.4012 | No |
| 79 | GRK6 | GRK6 Entrez, Source | G protein-coupled receptor kinase 6 | 6012 | 0.005 | 0.3995 | No |
| 80 | PRKACA | PRKACA Entrez, Source | protein kinase, cAMP-dependent, catalytic, alpha | 6017 | 0.005 | 0.3995 | No |
| 81 | KCNJ3 | KCNJ3 Entrez, Source | potassium inwardly-rectifying channel, subfamily J, member 3 | 6023 | 0.005 | 0.3995 | No |
| 82 | YWHAB | YWHAB Entrez, Source | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide | 6058 | 0.004 | 0.3972 | No |
| 83 | KCNJ5 | KCNJ5 Entrez, Source | potassium inwardly-rectifying channel, subfamily J, member 5 | 6397 | -0.002 | 0.3717 | No |
| 84 | GJB4 | GJB4 Entrez, Source | gap junction protein, beta 4 (connexin 30.3) | 6443 | -0.002 | 0.3685 | No |
| 85 | GNAO1 | GNAO1 Entrez, Source | guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O | 6475 | -0.003 | 0.3664 | No |
| 86 | CACNA1C | CACNA1C Entrez, Source | calcium channel, voltage-dependent, L type, alpha 1C subunit | 6752 | -0.008 | 0.3461 | No |
| 87 | NME7 | NME7 Entrez, Source | non-metastatic cells 7, protein expressed in (nucleoside-diphosphate kinase) | 6771 | -0.008 | 0.3453 | No |
| 88 | ADRA1B | ADRA1B Entrez, Source | adrenergic, alpha-1B-, receptor | 6830 | -0.009 | 0.3416 | No |
| 89 | CALM2 | CALM2 Entrez, Source | calmodulin 2 (phosphorylase kinase, delta) | 6851 | -0.010 | 0.3407 | No |
| 90 | GNAZ | GNAZ Entrez, Source | guanine nucleotide binding protein (G protein), alpha z polypeptide | 7190 | -0.016 | 0.3163 | No |
| 91 | GNG5 | GNG5 Entrez, Source | guanine nucleotide binding protein (G protein), gamma 5 | 7340 | -0.018 | 0.3063 | No |
| 92 | YWHAH | YWHAH Entrez, Source | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide | 7564 | -0.023 | 0.2911 | No |
| 93 | CAMK2G | CAMK2G Entrez, Source | calcium/calmodulin-dependent protein kinase (CaM kinase) II gamma | 7620 | -0.024 | 0.2886 | No |
| 94 | GJA5 | GJA5 Entrez, Source | gap junction protein, alpha 5, 40kDa (connexin 40) | 7650 | -0.025 | 0.2881 | No |
| 95 | PRKCD | PRKCD Entrez, Source | protein kinase C, delta | 7739 | -0.026 | 0.2833 | No |
| 96 | PRKCQ | PRKCQ Entrez, Source | protein kinase C, theta | 7952 | -0.030 | 0.2694 | No |
| 97 | CACNA1A | CACNA1A Entrez, Source | calcium channel, voltage-dependent, P/Q type, alpha 1A subunit | 8022 | -0.032 | 0.2664 | No |
| 98 | RGS14 | RGS14 Entrez, Source | regulator of G-protein signalling 14 | 8231 | -0.036 | 0.2533 | No |
| 99 | RYR2 | RYR2 Entrez, Source | ryanodine receptor 2 (cardiac) | 8394 | -0.039 | 0.2438 | No |
| 100 | CAMK2B | CAMK2B Entrez, Source | calcium/calmodulin-dependent protein kinase (CaM kinase) II beta | 8471 | -0.041 | 0.2409 | No |
| 101 | ADCY3 | ADCY3 Entrez, Source | adenylate cyclase 3 | 8509 | -0.042 | 0.2411 | No |
| 102 | ATP2A2 | ATP2A2 Entrez, Source | ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 | 8616 | -0.044 | 0.2361 | No |
| 103 | CACNA1S | CACNA1S Entrez, Source | calcium channel, voltage-dependent, L type, alpha 1S subunit | 8978 | -0.051 | 0.2125 | No |
| 104 | CHRM1 | CHRM1 Entrez, Source | cholinergic receptor, muscarinic 1 | 9204 | -0.056 | 0.1994 | No |
| 105 | CASQ2 | CASQ2 Entrez, Source | calsequestrin 2 (cardiac muscle) | 9360 | -0.060 | 0.1919 | No |
| 106 | GNA11 | GNA11 Entrez, Source | guanine nucleotide binding protein (G protein), alpha 11 (Gq class) | 9547 | -0.064 | 0.1823 | No |
| 107 | GNG12 | GNG12 Entrez, Source | guanine nucleotide binding protein (G protein), gamma 12 | 9591 | -0.065 | 0.1837 | No |
| 108 | SLC8A3 | SLC8A3 Entrez, Source | solute carrier family 8 (sodium-calcium exchanger), member 3 | 9596 | -0.065 | 0.1880 | No |
| 109 | ATP2B1 | ATP2B1 Entrez, Source | ATPase, Ca++ transporting, plasma membrane 1 | 9817 | -0.071 | 0.1764 | No |
| 110 | ADCY8 | ADCY8 Entrez, Source | adenylate cyclase 8 (brain) | 9950 | -0.075 | 0.1717 | No |
| 111 | ATP2B2 | ATP2B2 Entrez, Source | ATPase, Ca++ transporting, plasma membrane 2 | 9983 | -0.076 | 0.1746 | No |
| 112 | ADRA1A | ADRA1A Entrez, Source | adrenergic, alpha-1A-, receptor | 10025 | -0.077 | 0.1770 | No |
| 113 | ADRB1 | ADRB1 Entrez, Source | adrenergic, beta-1-, receptor | 10250 | -0.083 | 0.1659 | No |
| 114 | PRKCE | PRKCE Entrez, Source | protein kinase C, epsilon | 10468 | -0.089 | 0.1558 | No |
| 115 | ADCY4 | ADCY4 Entrez, Source | adenylate cyclase 4 | 10641 | -0.095 | 0.1495 | No |
| 116 | CALM1 | CALM1 Entrez, Source | calmodulin 1 (phosphorylase kinase, delta) | 10674 | -0.096 | 0.1539 | No |
| 117 | PRKD1 | PRKD1 Entrez, Source | protein kinase D1 | 11159 | -0.115 | 0.1254 | No |
| 118 | CALR | CALR Entrez, Source | calreticulin | 11620 | -0.135 | 0.1001 | No |
| 119 | PRKCB1 | PRKCB1 Entrez, Source | protein kinase C, beta 1 | 11650 | -0.136 | 0.1075 | No |
| 120 | ATP1B3 | ATP1B3 Entrez, Source | ATPase, Na+/K+ transporting, beta 3 polypeptide | 11707 | -0.138 | 0.1130 | No |
| 121 | GNB1 | GNB1 Entrez, Source | guanine nucleotide binding protein (G protein), beta polypeptide 1 | 11879 | -0.149 | 0.1106 | No |