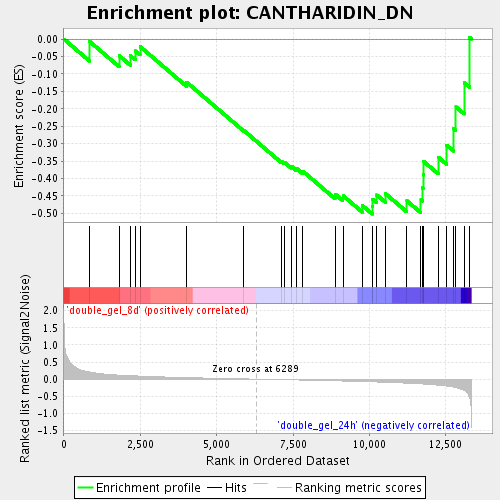
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_double_gel_24h.class.cls #double_gel_8d_versus_double_gel_24h_repos |
| Phenotype | class.cls#double_gel_8d_versus_double_gel_24h_repos |
| Upregulated in class | double_gel_24h |
| GeneSet | CANTHARIDIN_DN |
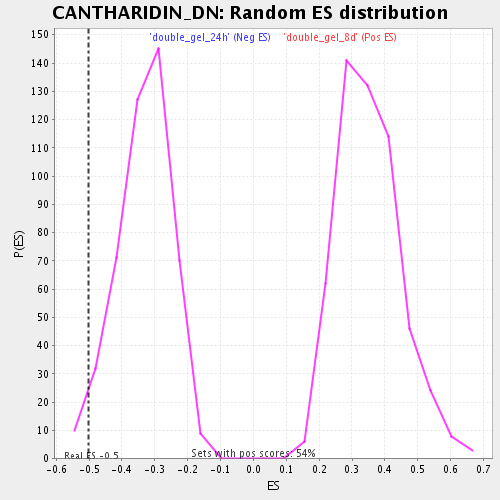
| Enrichment Score (ES) | -0.501924 |
| Normalized Enrichment Score (NES) | -1.5072833 |
| Nominal p-value | 0.030172413 |
| FDR q-value | 0.22747414 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | IDH3A | IDH3A Entrez, Source | isocitrate dehydrogenase 3 (NAD+) alpha | 843 | 0.206 | -0.0070 | No |
| 2 | EML2 | EML2 Entrez, Source | echinoderm microtubule associated protein like 2 | 1808 | 0.117 | -0.0475 | No |
| 3 | HADHA | HADHA Entrez, Source | hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (trifunctional protein), alpha subunit | 2173 | 0.101 | -0.0473 | No |
| 4 | ABCA3 | ABCA3 Entrez, Source | ATP-binding cassette, sub-family A (ABC1), member 3 | 2338 | 0.094 | -0.0339 | No |
| 5 | UQCRH | UQCRH Entrez, Source | ubiquinol-cytochrome c reductase hinge protein | 2494 | 0.088 | -0.0215 | No |
| 6 | ATP5G3 | ATP5G3 Entrez, Source | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C3 (subunit 9) | 4011 | 0.044 | -0.1233 | No |
| 7 | PFKL | PFKL Entrez, Source | phosphofructokinase, liver | 5889 | 0.007 | -0.2623 | No |
| 8 | TRAP1 | TRAP1 Entrez, Source | TNF receptor-associated protein 1 | 7108 | -0.014 | -0.3499 | No |
| 9 | FARSLB | FARSLB Entrez, Source | phenylalanine-tRNA synthetase-like, beta subunit | 7214 | -0.016 | -0.3533 | No |
| 10 | QDPR | QDPR Entrez, Source | quinoid dihydropteridine reductase | 7441 | -0.021 | -0.3647 | No |
| 11 | MTHFD1 | MTHFD1 Entrez, Source | methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase | 7607 | -0.024 | -0.3706 | No |
| 12 | SLK | SLK Entrez, Source | STE20-like kinase (yeast) | 7815 | -0.028 | -0.3786 | No |
| 13 | RPL29 | RPL29 Entrez, Source | ribosomal protein L29 | 8874 | -0.049 | -0.4446 | No |
| 14 | PIK3CA | PIK3CA Entrez, Source | phosphoinositide-3-kinase, catalytic, alpha polypeptide | 9133 | -0.055 | -0.4490 | No |
| 15 | RPL15 | RPL15 Entrez, Source | ribosomal protein L15 | 9763 | -0.070 | -0.4772 | No |
| 16 | PGAM1 | PGAM1 Entrez, Source | phosphoglycerate mutase 1 (brain) | 10093 | -0.079 | -0.4803 | Yes |
| 17 | GTF3A | GTF3A Entrez, Source | general transcription factor IIIA | 10103 | -0.079 | -0.4593 | Yes |
| 18 | RPL4 | RPL4 Entrez, Source | ribosomal protein L4 | 10242 | -0.083 | -0.4470 | Yes |
| 19 | POLD2 | POLD2 Entrez, Source | polymerase (DNA directed), delta 2, regulatory subunit 50kDa | 10527 | -0.091 | -0.4435 | Yes |
| 20 | FABP5 | FABP5 Entrez, Source | fatty acid binding protein 5 (psoriasis-associated) | 11225 | -0.117 | -0.4639 | Yes |
| 21 | RUVBL2 | RUVBL2 Entrez, Source | RuvB-like 2 (E. coli) | 11685 | -0.137 | -0.4608 | Yes |
| 22 | EIF3S9 | EIF3S9 Entrez, Source | eukaryotic translation initiation factor 3, subunit 9 eta, 116kDa | 11737 | -0.140 | -0.4262 | Yes |
| 23 | EEF1D | EEF1D Entrez, Source | eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) | 11758 | -0.142 | -0.3890 | Yes |
| 24 | HNRPC | HNRPC Entrez, Source | heterogeneous nuclear ribonucleoprotein C (C1/C2) | 11772 | -0.143 | -0.3509 | Yes |
| 25 | MYC | MYC Entrez, Source | v-myc myelocytomatosis viral oncogene homolog (avian) | 12249 | -0.175 | -0.3386 | Yes |
| 26 | TKT | TKT Entrez, Source | transketolase (Wernicke-Korsakoff syndrome) | 12523 | -0.202 | -0.3039 | Yes |
| 27 | SNRPA | SNRPA Entrez, Source | small nuclear ribonucleoprotein polypeptide A | 12756 | -0.234 | -0.2573 | Yes |
| 28 | PRIM1 | PRIM1 Entrez, Source | primase, polypeptide 1, 49kDa | 12830 | -0.249 | -0.1946 | Yes |
| 29 | MCM7 | MCM7 Entrez, Source | MCM7 minichromosome maintenance deficient 7 (S. cerevisiae) | 13101 | -0.328 | -0.1252 | Yes |
| 30 | ABCC4 | ABCC4 Entrez, Source | ATP-binding cassette, sub-family C (CFTR/MRP), member 4 | 13272 | -0.523 | 0.0053 | Yes |