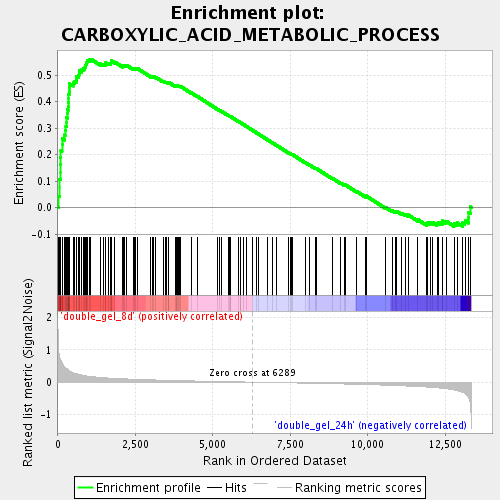
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_double_gel_24h.class.cls #double_gel_8d_versus_double_gel_24h_repos |
| Phenotype | class.cls#double_gel_8d_versus_double_gel_24h_repos |
| Upregulated in class | double_gel_8d |
| GeneSet | CARBOXYLIC_ACID_METABOLIC_PROCESS |
| Enrichment Score (ES) | 0.5599325 |
| Normalized Enrichment Score (NES) | 2.080857 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0019684639 |
| FWER p-Value | 0.03 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | HAO2 | HAO2 Entrez, Source | hydroxyacid oxidase 2 (long chain) | 18 | 1.071 | 0.0420 | Yes |
| 2 | FTCD | FTCD Entrez, Source | formiminotransferase cyclodeaminase | 46 | 0.834 | 0.0738 | Yes |
| 3 | PFKFB1 | PFKFB1 Entrez, Source | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 | 47 | 0.829 | 0.1073 | Yes |
| 4 | GOT1 | GOT1 Entrez, Source | glutamic-oxaloacetic transaminase 1, soluble (aspartate aminotransferase 1) | 76 | 0.697 | 0.1334 | Yes |
| 5 | BBOX1 | BBOX1 Entrez, Source | butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 | 78 | 0.695 | 0.1615 | Yes |
| 6 | CPT1A | CPT1A Entrez, Source | carnitine palmitoyltransferase 1A (liver) | 84 | 0.686 | 0.1889 | Yes |
| 7 | ACOX2 | ACOX2 Entrez, Source | acyl-Coenzyme A oxidase 2, branched chain | 97 | 0.667 | 0.2150 | Yes |
| 8 | PAH | PAH Entrez, Source | phenylalanine hydroxylase | 133 | 0.597 | 0.2365 | Yes |
| 9 | HGD | HGD Entrez, Source | homogentisate 1,2-dioxygenase (homogentisate oxidase) | 138 | 0.591 | 0.2601 | Yes |
| 10 | AGXT | AGXT Entrez, Source | alanine-glyoxylate aminotransferase (oxalosis I; hyperoxaluria I; glycolicaciduria; serine-pyruvate aminotransferase) | 212 | 0.475 | 0.2738 | Yes |
| 11 | FAH | FAH Entrez, Source | fumarylacetoacetate hydrolase (fumarylacetoacetase) | 232 | 0.457 | 0.2909 | Yes |
| 12 | BCKDHA | BCKDHA Entrez, Source | branched chain keto acid dehydrogenase E1, alpha polypeptide | 258 | 0.431 | 0.3065 | Yes |
| 13 | GGT1 | GGT1 Entrez, Source | gamma-glutamyltransferase 1 | 271 | 0.423 | 0.3227 | Yes |
| 14 | DDAH1 | DDAH1 Entrez, Source | dimethylarginine dimethylaminohydrolase 1 | 276 | 0.416 | 0.3392 | Yes |
| 15 | ALDH6A1 | ALDH6A1 Entrez, Source | aldehyde dehydrogenase 6 family, member A1 | 300 | 0.402 | 0.3538 | Yes |
| 16 | ARG1 | ARG1 Entrez, Source | arginase, liver | 307 | 0.397 | 0.3694 | Yes |
| 17 | FADS1 | FADS1 Entrez, Source | fatty acid desaturase 1 | 330 | 0.373 | 0.3828 | Yes |
| 18 | SLC25A15 | SLC25A15 Entrez, Source | solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 15 | 338 | 0.371 | 0.3973 | Yes |
| 19 | ECHS1 | ECHS1 Entrez, Source | enoyl Coenzyme A hydratase, short chain, 1, mitochondrial | 346 | 0.367 | 0.4116 | Yes |
| 20 | ECH1 | ECH1 Entrez, Source | enoyl Coenzyme A hydratase 1, peroxisomal | 350 | 0.365 | 0.4262 | Yes |
| 21 | MAT1A | MAT1A Entrez, Source | methionine adenosyltransferase I, alpha | 362 | 0.357 | 0.4398 | Yes |
| 22 | SLC3A1 | SLC3A1 Entrez, Source | 1 | 373 | 0.349 | 0.4532 | Yes |
| 23 | GLYAT | GLYAT Entrez, Source | glycine-N-acyltransferase | 379 | 0.348 | 0.4669 | Yes |
| 24 | HACL1 | HACL1 Entrez, Source | 2-hydroxyacyl-CoA lyase 1 | 490 | 0.289 | 0.4703 | Yes |
| 25 | GLS2 | GLS2 Entrez, Source | glutaminase 2 (liver, mitochondrial) | 543 | 0.271 | 0.4773 | Yes |
| 26 | ALDH5A1 | ALDH5A1 Entrez, Source | aldehyde dehydrogenase 5 family, member A1 (succinate-semialdehyde dehydrogenase) | 595 | 0.258 | 0.4839 | Yes |
| 27 | BAAT | BAAT Entrez, Source | bile acid Coenzyme A: amino acid N-acyltransferase (glycine N-choloyltransferase) | 602 | 0.257 | 0.4938 | Yes |
| 28 | MCCC2 | MCCC2 Entrez, Source | methylcrotonoyl-Coenzyme A carboxylase 2 (beta) | 663 | 0.243 | 0.4991 | Yes |
| 29 | ADI1 | ADI1 Entrez, Source | acireductone dioxygenase 1 | 682 | 0.239 | 0.5074 | Yes |
| 30 | CDO1 | CDO1 Entrez, Source | cysteine dioxygenase, type I | 689 | 0.237 | 0.5166 | Yes |
| 31 | AS3MT | AS3MT Entrez, Source | arsenic (+3 oxidation state) methyltransferase | 771 | 0.221 | 0.5194 | Yes |
| 32 | MLYCD | MLYCD Entrez, Source | malonyl-CoA decarboxylase | 817 | 0.210 | 0.5245 | Yes |
| 33 | BCKDK | BCKDK Entrez, Source | branched chain ketoacid dehydrogenase kinase | 869 | 0.200 | 0.5287 | Yes |
| 34 | PECI | PECI Entrez, Source | peroxisomal D3,D2-enoyl-CoA isomerase | 886 | 0.197 | 0.5355 | Yes |
| 35 | BCKDHB | BCKDHB Entrez, Source | branched chain keto acid dehydrogenase E1, beta polypeptide (maple syrup urine disease) | 906 | 0.195 | 0.5420 | Yes |
| 36 | FADS2 | FADS2 Entrez, Source | fatty acid desaturase 2 | 934 | 0.190 | 0.5476 | Yes |
| 37 | ACO2 | ACO2 Entrez, Source | aconitase 2, mitochondrial | 940 | 0.189 | 0.5549 | Yes |
| 38 | SDS | SDS Entrez, Source | serine dehydratase | 1007 | 0.182 | 0.5573 | Yes |
| 39 | PPARGC1A | PPARGC1A Entrez, Source | peroxisome proliferative activated receptor, gamma, coactivator 1, alpha | 1066 | 0.174 | 0.5599 | Yes |
| 40 | MSRA | MSRA Entrez, Source | methionine sulfoxide reductase A | 1379 | 0.144 | 0.5422 | No |
| 41 | PTS | PTS Entrez, Source | 6-pyruvoyltetrahydropterin synthase | 1478 | 0.137 | 0.5403 | No |
| 42 | GAMT | GAMT Entrez, Source | guanidinoacetate N-methyltransferase | 1535 | 0.132 | 0.5414 | No |
| 43 | ACADS | ACADS Entrez, Source | acyl-Coenzyme A dehydrogenase, C-2 to C-3 short chain | 1544 | 0.132 | 0.5461 | No |
| 44 | ME1 | ME1 Entrez, Source | malic enzyme 1, NADP(+)-dependent, cytosolic | 1647 | 0.125 | 0.5435 | No |
| 45 | HPGD | HPGD Entrez, Source | hydroxyprostaglandin dehydrogenase 15-(NAD) | 1689 | 0.123 | 0.5454 | No |
| 46 | SLC7A2 | SLC7A2 Entrez, Source | solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 | 1713 | 0.122 | 0.5486 | No |
| 47 | ACADSB | ACADSB Entrez, Source | acyl-Coenzyme A dehydrogenase, short/branched chain | 1715 | 0.122 | 0.5534 | No |
| 48 | DDAH2 | DDAH2 Entrez, Source | dimethylarginine dimethylaminohydrolase 2 | 1832 | 0.115 | 0.5493 | No |
| 49 | ASPA | ASPA Entrez, Source | aspartoacylase (Canavan disease) | 2090 | 0.104 | 0.5341 | No |
| 50 | AKR1D1 | AKR1D1 Entrez, Source | aldo-keto reductase family 1, member D1 (delta 4-3-ketosteroid-5-beta-reductase) | 2111 | 0.103 | 0.5367 | No |
| 51 | HDC | HDC Entrez, Source | histidine decarboxylase | 2155 | 0.101 | 0.5376 | No |
| 52 | AMT | AMT Entrez, Source | aminomethyltransferase | 2221 | 0.098 | 0.5367 | No |
| 53 | LYPLA3 | LYPLA3 Entrez, Source | lysophospholipase 3 (lysosomal phospholipase A2) | 2431 | 0.090 | 0.5245 | No |
| 54 | ACOT12 | ACOT12 Entrez, Source | acyl-CoA thioesterase 12 | 2485 | 0.088 | 0.5241 | No |
| 55 | AGPAT1 | AGPAT1 Entrez, Source | 1-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha) | 2510 | 0.088 | 0.5258 | No |
| 56 | NFS1 | NFS1 Entrez, Source | NFS1 nitrogen fixation 1 (S. cerevisiae) | 2570 | 0.085 | 0.5247 | No |
| 57 | ACADVL | ACADVL Entrez, Source | acyl-Coenzyme A dehydrogenase, very long chain | 3002 | 0.071 | 0.4950 | No |
| 58 | GNE | GNE Entrez, Source | glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase | 3063 | 0.069 | 0.4933 | No |
| 59 | FAAH | FAAH Entrez, Source | fatty acid amide hydrolase | 3091 | 0.069 | 0.4940 | No |
| 60 | IDH1 | IDH1 Entrez, Source | isocitrate dehydrogenase 1 (NADP+), soluble | 3161 | 0.067 | 0.4915 | No |
| 61 | ADIPOR1 | ADIPOR1 Entrez, Source | adiponectin receptor 1 | 3394 | 0.060 | 0.4764 | No |
| 62 | ACO1 | ACO1 Entrez, Source | aconitase 1, soluble | 3462 | 0.059 | 0.4737 | No |
| 63 | LTA4H | LTA4H Entrez, Source | leukotriene A4 hydrolase | 3505 | 0.057 | 0.4728 | No |
| 64 | SLC7A9 | SLC7A9 Entrez, Source | solute carrier family 7 (cationic amino acid transporter, y+ system), member 9 | 3559 | 0.056 | 0.4711 | No |
| 65 | DEGS1 | DEGS1 Entrez, Source | degenerative spermatocyte homolog 1, lipid desaturase (Drosophila) | 3581 | 0.055 | 0.4717 | No |
| 66 | ACADM | ACADM Entrez, Source | acyl-Coenzyme A dehydrogenase, C-4 to C-12 straight chain | 3794 | 0.050 | 0.4577 | No |
| 67 | FARS2 | FARS2 Entrez, Source | phenylalanine-tRNA synthetase 2 (mitochondrial) | 3826 | 0.050 | 0.4574 | No |
| 68 | SCLY | SCLY Entrez, Source | selenocysteine lyase | 3833 | 0.049 | 0.4589 | No |
| 69 | ACLY | ACLY Entrez, Source | ATP citrate lyase | 3867 | 0.048 | 0.4584 | No |
| 70 | GCSH | GCSH Entrez, Source | glycine cleavage system protein H (aminomethyl carrier) | 3887 | 0.048 | 0.4589 | No |
| 71 | BCAT1 | BCAT1 Entrez, Source | branched chain aminotransferase 1, cytosolic | 3938 | 0.046 | 0.4569 | No |
| 72 | SLC25A10 | SLC25A10 Entrez, Source | solute carrier family 25 (mitochondrial carrier; dicarboxylate transporter), member 10 | 3966 | 0.045 | 0.4567 | No |
| 73 | GLUD1 | GLUD1 Entrez, Source | glutamate dehydrogenase 1 | 4297 | 0.038 | 0.4333 | No |
| 74 | GBA2 | GBA2 Entrez, Source | glucosidase, beta (bile acid) 2 | 4489 | 0.034 | 0.4202 | No |
| 75 | ACOT2 | ACOT2 Entrez, Source | acyl-CoA thioesterase 2 | 5165 | 0.021 | 0.3700 | No |
| 76 | ABCD2 | ABCD2 Entrez, Source | ATP-binding cassette, sub-family D (ALD), member 2 | 5216 | 0.020 | 0.3670 | No |
| 77 | CPT1B | CPT1B Entrez, Source | carnitine palmitoyltransferase 1B (muscle) | 5265 | 0.019 | 0.3641 | No |
| 78 | ADIPOR2 | ADIPOR2 Entrez, Source | adiponectin receptor 2 | 5506 | 0.014 | 0.3466 | No |
| 79 | PPARD | PPARD Entrez, Source | peroxisome proliferative activated receptor, delta | 5532 | 0.014 | 0.3452 | No |
| 80 | PTGS1 | PTGS1 Entrez, Source | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | 5556 | 0.014 | 0.3440 | No |
| 81 | GGTLA1 | GGTLA1 Entrez, Source | gamma-glutamyltransferase-like activity 1 | 5579 | 0.013 | 0.3429 | No |
| 82 | SLC7A8 | SLC7A8 Entrez, Source | solute carrier family 7 (cationic amino acid transporter, y+ system), member 8 | 5837 | 0.008 | 0.3238 | No |
| 83 | SC4MOL | SC4MOL Entrez, Source | sterol-C4-methyl oxidase-like | 5880 | 0.008 | 0.3209 | No |
| 84 | PTGDS | PTGDS Entrez, Source | prostaglandin D2 synthase 21kDa (brain) | 5900 | 0.007 | 0.3198 | No |
| 85 | ALDH1L1 | ALDH1L1 Entrez, Source | aldehyde dehydrogenase 1 family, member L1 | 5998 | 0.005 | 0.3126 | No |
| 86 | ASRGL1 | ASRGL1 Entrez, Source | asparaginase like 1 | 6099 | 0.003 | 0.3052 | No |
| 87 | ATF4 | ATF4 Entrez, Source | activating transcription factor 4 (tax-responsive enhancer element B67) | 6295 | -0.000 | 0.2905 | No |
| 88 | PTGES | PTGES Entrez, Source | prostaglandin E synthase | 6415 | -0.002 | 0.2815 | No |
| 89 | HPD | HPD Entrez, Source | 4-hydroxyphenylpyruvate dioxygenase | 6481 | -0.003 | 0.2767 | No |
| 90 | ASL | ASL Entrez, Source | argininosuccinate lyase | 6773 | -0.008 | 0.2551 | No |
| 91 | SLC6A6 | SLC6A6 Entrez, Source | solute carrier family 6 (neurotransmitter transporter, taurine), member 6 | 6923 | -0.011 | 0.2442 | No |
| 92 | CYP7B1 | CYP7B1 Entrez, Source | cytochrome P450, family 7, subfamily B, polypeptide 1 | 7069 | -0.014 | 0.2338 | No |
| 93 | QDPR | QDPR Entrez, Source | quinoid dihydropteridine reductase | 7441 | -0.021 | 0.2065 | No |
| 94 | ACN9 | ACN9 Entrez, Source | ACN9 homolog (S. cerevisiae) | 7494 | -0.022 | 0.2035 | No |
| 95 | GSS | GSS Entrez, Source | glutathione synthetase | 7538 | -0.022 | 0.2011 | No |
| 96 | NANP | NANP Entrez, Source | N-acetylneuraminic acid phosphatase | 7570 | -0.023 | 0.1997 | No |
| 97 | HADHB | HADHB Entrez, Source | hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (trifunctional protein), beta subunit | 7978 | -0.031 | 0.1701 | No |
| 98 | CYP4F2 | CYP4F2 Entrez, Source | cytochrome P450, family 4, subfamily F, polypeptide 2 | 8129 | -0.034 | 0.1602 | No |
| 99 | TNFRSF1A | TNFRSF1A Entrez, Source | tumor necrosis factor receptor superfamily, member 1A | 8306 | -0.037 | 0.1484 | No |
| 100 | HSD3B7 | HSD3B7 Entrez, Source | hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 | 8340 | -0.038 | 0.1474 | No |
| 101 | IDH3B | IDH3B Entrez, Source | isocitrate dehydrogenase 3 (NAD+) beta | 8861 | -0.049 | 0.1100 | No |
| 102 | IGF1 | IGF1 Entrez, Source | insulin-like growth factor 1 (somatomedin C) | 9134 | -0.055 | 0.0917 | No |
| 103 | ARPP-19 | ARPP-19 Entrez, Source | - | 9233 | -0.057 | 0.0865 | No |
| 104 | GAD1 | GAD1 Entrez, Source | glutamate decarboxylase 1 (brain, 67kDa) | 9270 | -0.057 | 0.0861 | No |
| 105 | GOT2 | GOT2 Entrez, Source | glutamic-oxaloacetic transaminase 2, mitochondrial (aspartate aminotransferase 2) | 9641 | -0.066 | 0.0608 | No |
| 106 | NR1H4 | NR1H4 Entrez, Source | nuclear receptor subfamily 1, group H, member 4 | 9939 | -0.075 | 0.0414 | No |
| 107 | YARS | YARS Entrez, Source | tyrosyl-tRNA synthetase | 9947 | -0.075 | 0.0439 | No |
| 108 | INDO | INDO Entrez, Source | indoleamine-pyrrole 2,3 dioxygenase | 10558 | -0.092 | 0.0014 | No |
| 109 | GATM | GATM Entrez, Source | glycine amidinotransferase (L-arginine:glycine amidinotransferase) | 10782 | -0.100 | -0.0114 | No |
| 110 | ACOX3 | ACOX3 Entrez, Source | acyl-Coenzyme A oxidase 3, pristanoyl | 10884 | -0.104 | -0.0148 | No |
| 111 | KARS | KARS Entrez, Source | lysyl-tRNA synthetase | 10927 | -0.106 | -0.0137 | No |
| 112 | BRCA1 | BRCA1 Entrez, Source | breast cancer 1, early onset | 11096 | -0.112 | -0.0219 | No |
| 113 | GAD2 | GAD2 Entrez, Source | glutamate decarboxylase 2 (pancreatic islets and brain, 65kDa) | 11204 | -0.116 | -0.0253 | No |
| 114 | PLOD1 | PLOD1 Entrez, Source | procollagen-lysine 1, 2-oxoglutarate 5-dioxygenase 1 | 11305 | -0.121 | -0.0280 | No |
| 115 | GNPAT | GNPAT Entrez, Source | glyceronephosphate O-acyltransferase | 11617 | -0.134 | -0.0461 | No |
| 116 | SLC7A7 | SLC7A7 Entrez, Source | solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 | 11894 | -0.150 | -0.0609 | No |
| 117 | CROT | CROT Entrez, Source | carnitine O-octanoyltransferase | 11938 | -0.152 | -0.0580 | No |
| 118 | DARS | DARS Entrez, Source | aspartyl-tRNA synthetase | 12016 | -0.158 | -0.0574 | No |
| 119 | WARS | WARS Entrez, Source | tryptophanyl-tRNA synthetase | 12088 | -0.163 | -0.0561 | No |
| 120 | AARS | AARS Entrez, Source | alanyl-tRNA synthetase | 12234 | -0.174 | -0.0601 | No |
| 121 | MIF | MIF Entrez, Source | macrophage migration inhibitory factor (glycosylation-inhibiting factor) | 12291 | -0.179 | -0.0571 | No |
| 122 | SCYE1 | SCYE1 Entrez, Source | small inducible cytokine subfamily E, member 1 (endothelial monocyte-activating) | 12401 | -0.188 | -0.0577 | No |
| 123 | PPARA | PPARA Entrez, Source | peroxisome proliferative activated receptor, alpha | 12405 | -0.189 | -0.0503 | No |
| 124 | SMS | SMS Entrez, Source | spermine synthase | 12527 | -0.202 | -0.0513 | No |
| 125 | GCLM | GCLM Entrez, Source | glutamate-cysteine ligase, modifier subunit | 12784 | -0.238 | -0.0610 | No |
| 126 | SLC7A5 | SLC7A5 Entrez, Source | solute carrier family 7 (cationic amino acid transporter, y+ system), member 5 | 12884 | -0.262 | -0.0579 | No |
| 127 | GCK | GCK Entrez, Source | glucokinase (hexokinase 4, maturity onset diabetes of the young 2) | 13052 | -0.307 | -0.0581 | No |
| 128 | UGDH | UGDH Entrez, Source | UDP-glucose dehydrogenase | 13138 | -0.352 | -0.0503 | No |
| 129 | GCLC | GCLC Entrez, Source | glutamate-cysteine ligase, catalytic subunit | 13249 | -0.480 | -0.0392 | No |
| 130 | SLC27A5 | SLC27A5 Entrez, Source | solute carrier family 27 (fatty acid transporter), member 5 | 13261 | -0.509 | -0.0194 | No |
| 131 | CD74 | CD74 Entrez, Source | CD74 molecule, major histocompatibility complex, class II invariant chain | 13307 | -0.629 | 0.0026 | No |

