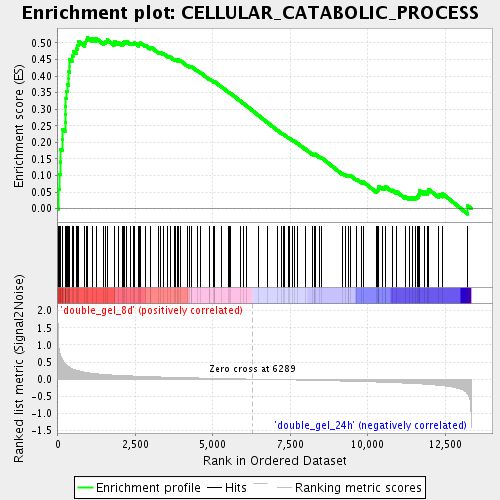
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | double_gel_and_single_gel_rma_expression_values_collapsed_to_symbols.class.cls #double_gel_8d_versus_double_gel_24h.class.cls #double_gel_8d_versus_double_gel_24h_repos |
| Phenotype | class.cls#double_gel_8d_versus_double_gel_24h_repos |
| Upregulated in class | double_gel_8d |
| GeneSet | CELLULAR_CATABOLIC_PROCESS |
| Enrichment Score (ES) | 0.5169039 |
| Normalized Enrichment Score (NES) | 1.9140096 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.011085371 |
| FWER p-Value | 0.466 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | HAO2 | HAO2 Entrez, Source | hydroxyacid oxidase 2 (long chain) | 18 | 1.071 | 0.0591 | Yes |
| 2 | PFKFB1 | PFKFB1 Entrez, Source | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 | 47 | 0.829 | 0.1038 | Yes |
| 3 | GOT1 | GOT1 Entrez, Source | glutamic-oxaloacetic transaminase 1, soluble (aspartate aminotransferase 1) | 76 | 0.697 | 0.1410 | Yes |
| 4 | CPT1A | CPT1A Entrez, Source | carnitine palmitoyltransferase 1A (liver) | 84 | 0.686 | 0.1792 | Yes |
| 5 | HGD | HGD Entrez, Source | homogentisate 1,2-dioxygenase (homogentisate oxidase) | 138 | 0.591 | 0.2086 | Yes |
| 6 | MIOX | MIOX Entrez, Source | myo-inositol oxygenase | 146 | 0.570 | 0.2402 | Yes |
| 7 | FAH | FAH Entrez, Source | fumarylacetoacetate hydrolase (fumarylacetoacetase) | 232 | 0.457 | 0.2596 | Yes |
| 8 | GPX3 | GPX3 Entrez, Source | glutathione peroxidase 3 (plasma) | 242 | 0.449 | 0.2843 | Yes |
| 9 | SMPD3 | SMPD3 Entrez, Source | sphingomyelin phosphodiesterase 3, neutral membrane (neutral sphingomyelinase II) | 248 | 0.442 | 0.3089 | Yes |
| 10 | BCKDHA | BCKDHA Entrez, Source | branched chain keto acid dehydrogenase E1, alpha polypeptide | 258 | 0.431 | 0.3326 | Yes |
| 11 | DDAH1 | DDAH1 Entrez, Source | dimethylarginine dimethylaminohydrolase 1 | 276 | 0.416 | 0.3547 | Yes |
| 12 | ARG1 | ARG1 Entrez, Source | arginase, liver | 307 | 0.397 | 0.3749 | Yes |
| 13 | ECHS1 | ECHS1 Entrez, Source | enoyl Coenzyme A hydratase, short chain, 1, mitochondrial | 346 | 0.367 | 0.3928 | Yes |
| 14 | ECH1 | ECH1 Entrez, Source | enoyl Coenzyme A hydratase 1, peroxisomal | 350 | 0.365 | 0.4131 | Yes |
| 15 | APOA4 | APOA4 Entrez, Source | apolipoprotein A-IV | 383 | 0.346 | 0.4302 | Yes |
| 16 | HK1 | HK1 Entrez, Source | hexokinase 1 | 385 | 0.345 | 0.4497 | Yes |
| 17 | AMPD3 | AMPD3 Entrez, Source | adenosine monophosphate deaminase (isoform E) | 458 | 0.303 | 0.4613 | Yes |
| 18 | HACL1 | HACL1 Entrez, Source | 2-hydroxyacyl-CoA lyase 1 | 490 | 0.289 | 0.4753 | Yes |
| 19 | ALDH5A1 | ALDH5A1 Entrez, Source | aldehyde dehydrogenase 5 family, member A1 (succinate-semialdehyde dehydrogenase) | 595 | 0.258 | 0.4820 | Yes |
| 20 | GUSB | GUSB Entrez, Source | glucuronidase, beta | 637 | 0.247 | 0.4928 | Yes |
| 21 | MCCC2 | MCCC2 Entrez, Source | methylcrotonoyl-Coenzyme A carboxylase 2 (beta) | 663 | 0.243 | 0.5047 | Yes |
| 22 | BCKDK | BCKDK Entrez, Source | branched chain ketoacid dehydrogenase kinase | 869 | 0.200 | 0.5004 | Yes |
| 23 | BCKDHB | BCKDHB Entrez, Source | branched chain keto acid dehydrogenase E1, beta polypeptide (maple syrup urine disease) | 906 | 0.195 | 0.5087 | Yes |
| 24 | ACO2 | ACO2 Entrez, Source | aconitase 2, mitochondrial | 940 | 0.189 | 0.5169 | Yes |
| 25 | STS | STS Entrez, Source | steroid sulfatase (microsomal), arylsulfatase C, isozyme S | 1105 | 0.170 | 0.5141 | No |
| 26 | RNASE6 | RNASE6 Entrez, Source | ribonuclease, RNase A family, k6 | 1232 | 0.158 | 0.5135 | No |
| 27 | APOA5 | APOA5 Entrez, Source | apolipoprotein A-V | 1486 | 0.136 | 0.5020 | No |
| 28 | ACADS | ACADS Entrez, Source | acyl-Coenzyme A dehydrogenase, C-2 to C-3 short chain | 1544 | 0.132 | 0.5052 | No |
| 29 | NUDT3 | NUDT3 Entrez, Source | nudix (nucleoside diphosphate linked moiety X)-type motif 3 | 1587 | 0.129 | 0.5093 | No |
| 30 | RNASEH2A | RNASEH2A Entrez, Source | ribonuclease H2, subunit A | 1815 | 0.116 | 0.4987 | No |
| 31 | DDAH2 | DDAH2 Entrez, Source | dimethylarginine dimethylaminohydrolase 2 | 1832 | 0.115 | 0.5039 | No |
| 32 | TST | TST Entrez, Source | thiosulfate sulfurtransferase (rhodanese) | 1969 | 0.109 | 0.4998 | No |
| 33 | ASPA | ASPA Entrez, Source | aspartoacylase (Canavan disease) | 2090 | 0.104 | 0.4966 | No |
| 34 | AKR1D1 | AKR1D1 Entrez, Source | aldo-keto reductase family 1, member D1 (delta 4-3-ketosteroid-5-beta-reductase) | 2111 | 0.103 | 0.5009 | No |
| 35 | ALDOB | ALDOB Entrez, Source | aldolase B, fructose-bisphosphate | 2152 | 0.101 | 0.5036 | No |
| 36 | AMT | AMT Entrez, Source | aminomethyltransferase | 2221 | 0.098 | 0.5040 | No |
| 37 | PPT1 | PPT1 Entrez, Source | palmitoyl-protein thioesterase 1 (ceroid-lipofuscinosis, neuronal 1, infantile) | 2353 | 0.093 | 0.4994 | No |
| 38 | LYPLA3 | LYPLA3 Entrez, Source | lysophospholipase 3 (lysosomal phospholipase A2) | 2431 | 0.090 | 0.4987 | No |
| 39 | ZFP36 | ZFP36 Entrez, Source | zinc finger protein 36, C3H type, homolog (mouse) | 2480 | 0.089 | 0.5000 | No |
| 40 | UBE4A | UBE4A Entrez, Source | ubiquitination factor E4A (UFD2 homolog, yeast) | 2612 | 0.083 | 0.4948 | No |
| 41 | GNS | GNS Entrez, Source | glucosamine (N-acetyl)-6-sulfatase (Sanfilippo disease IIID) | 2618 | 0.083 | 0.4991 | No |
| 42 | COLQ | COLQ Entrez, Source | collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase | 2670 | 0.081 | 0.4998 | No |
| 43 | SDHD | SDHD Entrez, Source | succinate dehydrogenase complex, subunit D, integral membrane protein | 2818 | 0.077 | 0.4931 | No |
| 44 | PYGB | PYGB Entrez, Source | phosphorylase, glycogen; brain | 2973 | 0.072 | 0.4855 | No |
| 45 | ACADVL | ACADVL Entrez, Source | acyl-Coenzyme A dehydrogenase, very long chain | 3002 | 0.071 | 0.4874 | No |
| 46 | CYCS | CYCS Entrez, Source | cytochrome c, somatic | 3256 | 0.064 | 0.4718 | No |
| 47 | SDHC | SDHC Entrez, Source | succinate dehydrogenase complex, subunit C, integral membrane protein, 15kDa | 3311 | 0.062 | 0.4713 | No |
| 48 | SIAH2 | SIAH2 Entrez, Source | seven in absentia homolog 2 (Drosophila) | 3412 | 0.060 | 0.4671 | No |
| 49 | GAA | GAA Entrez, Source | glucosidase, alpha; acid (Pompe disease, glycogen storage disease type II) | 3547 | 0.056 | 0.4601 | No |
| 50 | MPST | MPST Entrez, Source | mercaptopyruvate sulfurtransferase | 3621 | 0.054 | 0.4577 | No |
| 51 | PDHB | PDHB Entrez, Source | pyruvate dehydrogenase (lipoamide) beta | 3755 | 0.051 | 0.4505 | No |
| 52 | ACADM | ACADM Entrez, Source | acyl-Coenzyme A dehydrogenase, C-4 to C-12 straight chain | 3794 | 0.050 | 0.4504 | No |
| 53 | ACLY | ACLY Entrez, Source | ATP citrate lyase | 3867 | 0.048 | 0.4477 | No |
| 54 | GCSH | GCSH Entrez, Source | glycine cleavage system protein H (aminomethyl carrier) | 3887 | 0.048 | 0.4490 | No |
| 55 | UBE2B | UBE2B Entrez, Source | ubiquitin-conjugating enzyme E2B (RAD6 homolog) | 3964 | 0.045 | 0.4458 | No |
| 56 | PFKM | PFKM Entrez, Source | phosphofructokinase, muscle | 4179 | 0.040 | 0.4319 | No |
| 57 | SDHA | SDHA Entrez, Source | succinate dehydrogenase complex, subunit A, flavoprotein (Fp) | 4257 | 0.038 | 0.4282 | No |
| 58 | GLUD1 | GLUD1 Entrez, Source | glutamate dehydrogenase 1 | 4297 | 0.038 | 0.4274 | No |
| 59 | BTRC | BTRC Entrez, Source | beta-transducin repeat containing | 4316 | 0.037 | 0.4281 | No |
| 60 | GBA2 | GBA2 Entrez, Source | glucosidase, beta (bile acid) 2 | 4489 | 0.034 | 0.4171 | No |
| 61 | CDKN2A | CDKN2A Entrez, Source | cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4) | 4610 | 0.032 | 0.4098 | No |
| 62 | PRDX6 | PRDX6 Entrez, Source | peroxiredoxin 6 | 4883 | 0.026 | 0.3907 | No |
| 63 | NEU3 | NEU3 Entrez, Source | sialidase 3 (membrane sialidase) | 4899 | 0.026 | 0.3910 | No |
| 64 | DFFA | DFFA Entrez, Source | DNA fragmentation factor, 45kDa, alpha polypeptide | 5035 | 0.023 | 0.3821 | No |
| 65 | ALDOA | ALDOA Entrez, Source | aldolase A, fructose-bisphosphate | 5039 | 0.023 | 0.3832 | No |
| 66 | CEL | CEL Entrez, Source | carboxyl ester lipase (bile salt-stimulated lipase) | 5047 | 0.023 | 0.3840 | No |
| 67 | CPT1B | CPT1B Entrez, Source | carnitine palmitoyltransferase 1B (muscle) | 5265 | 0.019 | 0.3687 | No |
| 68 | TREH | TREH Entrez, Source | trehalase (brush-border membrane glycoprotein) | 5501 | 0.015 | 0.3517 | No |
| 69 | PPARD | PPARD Entrez, Source | peroxisome proliferative activated receptor, delta | 5532 | 0.014 | 0.3502 | No |
| 70 | UBE2N | UBE2N Entrez, Source | ubiquitin-conjugating enzyme E2N (UBC13 homolog, yeast) | 5570 | 0.013 | 0.3482 | No |
| 71 | PFKL | PFKL Entrez, Source | phosphofructokinase, liver | 5889 | 0.007 | 0.3245 | No |
| 72 | ALDH1L1 | ALDH1L1 Entrez, Source | aldehyde dehydrogenase 1 family, member L1 | 5998 | 0.005 | 0.3167 | No |
| 73 | ASRGL1 | ASRGL1 Entrez, Source | asparaginase like 1 | 6099 | 0.003 | 0.3093 | No |
| 74 | HPD | HPD Entrez, Source | 4-hydroxyphenylpyruvate dioxygenase | 6481 | -0.003 | 0.2806 | No |
| 75 | ASL | ASL Entrez, Source | argininosuccinate lyase | 6773 | -0.008 | 0.2591 | No |
| 76 | SOD1 | SOD1 Entrez, Source | superoxide dismutase 1, soluble (amyotrophic lateral sclerosis 1 (adult)) | 7093 | -0.014 | 0.2358 | No |
| 77 | STUB1 | STUB1 Entrez, Source | STIP1 homology and U-box containing protein 1 | 7226 | -0.016 | 0.2267 | No |
| 78 | UBE2D2 | UBE2D2 Entrez, Source | ubiquitin-conjugating enzyme E2D 2 (UBC4/5 homolog, yeast) | 7273 | -0.017 | 0.2242 | No |
| 79 | DHPS | DHPS Entrez, Source | deoxyhypusine synthase | 7301 | -0.018 | 0.2232 | No |
| 80 | NPLOC4 | NPLOC4 Entrez, Source | nuclear protein localization 4 homolog (S. cerevisiae) | 7452 | -0.021 | 0.2130 | No |
| 81 | ANAPC2 | ANAPC2 Entrez, Source | anaphase promoting complex subunit 2 | 7474 | -0.021 | 0.2126 | No |
| 82 | YWHAH | YWHAH Entrez, Source | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide | 7564 | -0.023 | 0.2072 | No |
| 83 | GAPDHS | GAPDHS Entrez, Source | glyceraldehyde-3-phosphate dehydrogenase, spermatogenic | 7619 | -0.024 | 0.2044 | No |
| 84 | PSMC5 | PSMC5 Entrez, Source | proteasome (prosome, macropain) 26S subunit, ATPase, 5 | 7735 | -0.026 | 0.1972 | No |
| 85 | HADHB | HADHB Entrez, Source | hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (trifunctional protein), beta subunit | 7978 | -0.031 | 0.1806 | No |
| 86 | UBB | UBB Entrez, Source | ubiquitin B | 8206 | -0.035 | 0.1654 | No |
| 87 | TPP1 | TPP1 Entrez, Source | tripeptidyl peptidase I | 8284 | -0.037 | 0.1617 | No |
| 88 | FBXO22 | FBXO22 Entrez, Source | F-box protein 22 | 8288 | -0.037 | 0.1636 | No |
| 89 | NEDD8 | NEDD8 Entrez, Source | neural precursor cell expressed, developmentally down-regulated 8 | 8297 | -0.037 | 0.1651 | No |
| 90 | UBE2I | UBE2I Entrez, Source | ubiquitin-conjugating enzyme E2I (UBC9 homolog, yeast) | 8446 | -0.040 | 0.1562 | No |
| 91 | ANAPC4 | ANAPC4 Entrez, Source | anaphase promoting complex subunit 4 | 8515 | -0.042 | 0.1534 | No |
| 92 | VCP | VCP Entrez, Source | valosin-containing protein | 9171 | -0.055 | 0.1070 | No |
| 93 | GAD1 | GAD1 Entrez, Source | glutamate decarboxylase 1 (brain, 67kDa) | 9270 | -0.057 | 0.1028 | No |
| 94 | CDC23 | CDC23 Entrez, Source | CDC23 (cell division cycle 23, yeast, homolog) | 9365 | -0.060 | 0.0991 | No |
| 95 | BLVRA | BLVRA Entrez, Source | biliverdin reductase A | 9379 | -0.060 | 0.1015 | No |
| 96 | DFFB | DFFB Entrez, Source | DNA fragmentation factor, 40kDa, beta polypeptide (caspase-activated DNase) | 9437 | -0.061 | 0.1006 | No |
| 97 | GOT2 | GOT2 Entrez, Source | glutamic-oxaloacetic transaminase 2, mitochondrial (aspartate aminotransferase 2) | 9641 | -0.066 | 0.0890 | No |
| 98 | UBE2D3 | UBE2D3 Entrez, Source | ubiquitin-conjugating enzyme E2D 3 (UBC4/5 homolog, yeast) | 9797 | -0.071 | 0.0813 | No |
| 99 | BNIP3 | BNIP3 Entrez, Source | BCL2/adenovirus E1B 19kDa interacting protein 3 | 9855 | -0.073 | 0.0811 | No |
| 100 | PABPC4 | PABPC4 Entrez, Source | poly(A) binding protein, cytoplasmic 4 (inducible form) | 10277 | -0.084 | 0.0539 | No |
| 101 | PARK2 | PARK2 Entrez, Source | Parkinson disease (autosomal recessive, juvenile) 2, parkin | 10307 | -0.085 | 0.0565 | No |
| 102 | SQSTM1 | SQSTM1 Entrez, Source | sequestosome 1 | 10340 | -0.086 | 0.0589 | No |
| 103 | ERCC3 | ERCC3 Entrez, Source | excision repair cross-complementing rodent repair deficiency, complementation group 3 (xeroderma pigmentosum group B complementing) | 10351 | -0.086 | 0.0630 | No |
| 104 | TSG101 | TSG101 Entrez, Source | tumor susceptibility gene 101 | 10354 | -0.086 | 0.0677 | No |
| 105 | BAX | BAX Entrez, Source | BCL2-associated X protein | 10485 | -0.090 | 0.0630 | No |
| 106 | INDO | INDO Entrez, Source | indoleamine-pyrrole 2,3 dioxygenase | 10558 | -0.092 | 0.0627 | No |
| 107 | GSPT1 | GSPT1 Entrez, Source | G1 to S phase transition 1 | 10578 | -0.093 | 0.0665 | No |
| 108 | UBE2G1 | UBE2G1 Entrez, Source | ubiquitin-conjugating enzyme E2G 1 (UBC7 homolog, yeast) | 10786 | -0.100 | 0.0565 | No |
| 109 | RNASEH1 | RNASEH1 Entrez, Source | ribonuclease H1 | 10932 | -0.106 | 0.0515 | No |
| 110 | GAD2 | GAD2 Entrez, Source | glutamate decarboxylase 2 (pancreatic islets and brain, 65kDa) | 11204 | -0.116 | 0.0376 | No |
| 111 | ABCE1 | ABCE1 Entrez, Source | ATP-binding cassette, sub-family E (OABP), member 1 | 11335 | -0.122 | 0.0346 | No |
| 112 | CDC20 | CDC20 Entrez, Source | CDC20 cell division cycle 20 homolog (S. cerevisiae) | 11434 | -0.126 | 0.0344 | No |
| 113 | UBE2V2 | UBE2V2 Entrez, Source | ubiquitin-conjugating enzyme E2 variant 2 | 11529 | -0.130 | 0.0346 | No |
| 114 | SYVN1 | SYVN1 Entrez, Source | synovial apoptosis inhibitor 1, synoviolin | 11594 | -0.133 | 0.0373 | No |
| 115 | UBE2A | UBE2A Entrez, Source | ubiquitin-conjugating enzyme E2A (RAD6 homolog) | 11635 | -0.135 | 0.0419 | No |
| 116 | FBXO7 | FBXO7 Entrez, Source | F-box protein 7 | 11665 | -0.136 | 0.0474 | No |
| 117 | USP33 | USP33 Entrez, Source | ubiquitin specific peptidase 33 | 11679 | -0.137 | 0.0541 | No |
| 118 | NUDT5 | NUDT5 Entrez, Source | nudix (nucleoside diphosphate linked moiety X)-type motif 5 | 11827 | -0.146 | 0.0513 | No |
| 119 | ISG20 | ISG20 Entrez, Source | interferon stimulated exonuclease gene 20kDa | 11943 | -0.153 | 0.0512 | No |
| 120 | UFD1L | UFD1L Entrez, Source | ubiquitin fusion degradation 1 like (yeast) | 11964 | -0.154 | 0.0584 | No |
| 121 | ARIH1 | ARIH1 Entrez, Source | ariadne homolog, ubiquitin-conjugating enzyme E2 binding protein, 1 (Drosophila) | 12297 | -0.179 | 0.0434 | No |
| 122 | PPARA | PPARA Entrez, Source | peroxisome proliferative activated receptor, alpha | 12405 | -0.189 | 0.0460 | No |
| 123 | HNRPD | HNRPD Entrez, Source | heterogeneous nuclear ribonucleoprotein D (AU-rich element RNA binding protein 1, 37kDa) | 13227 | -0.440 | 0.0087 | No |